Abstract
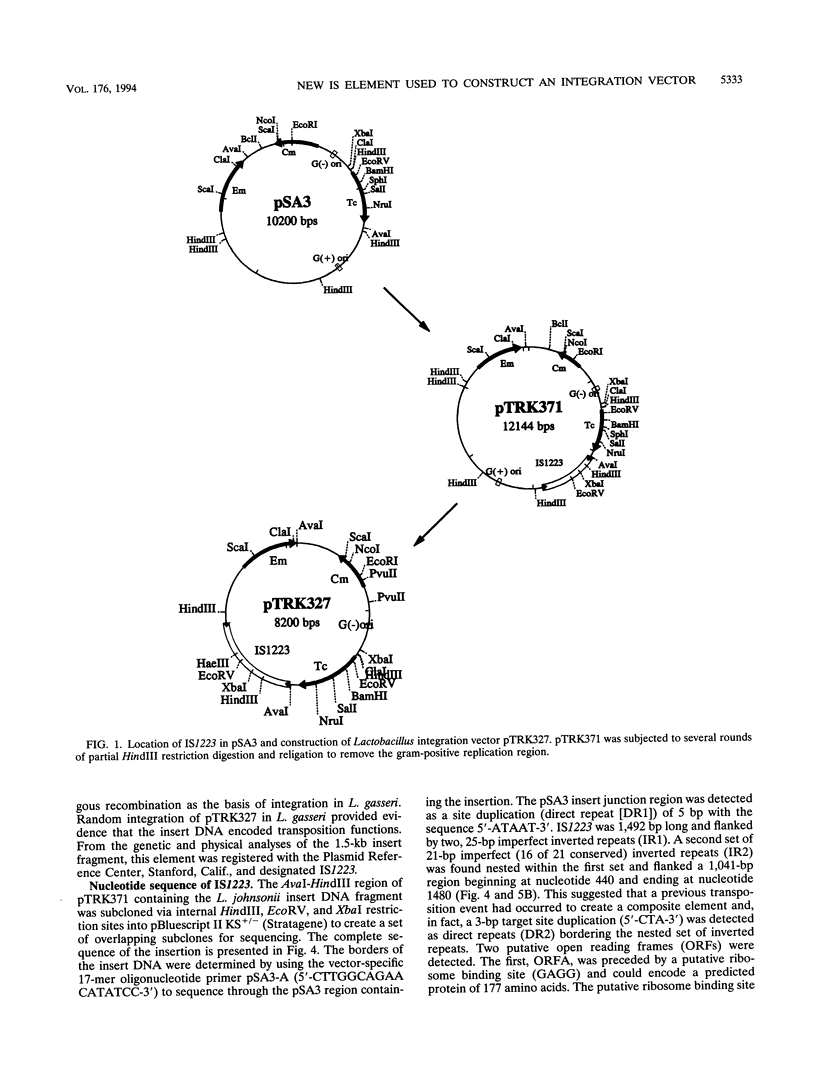
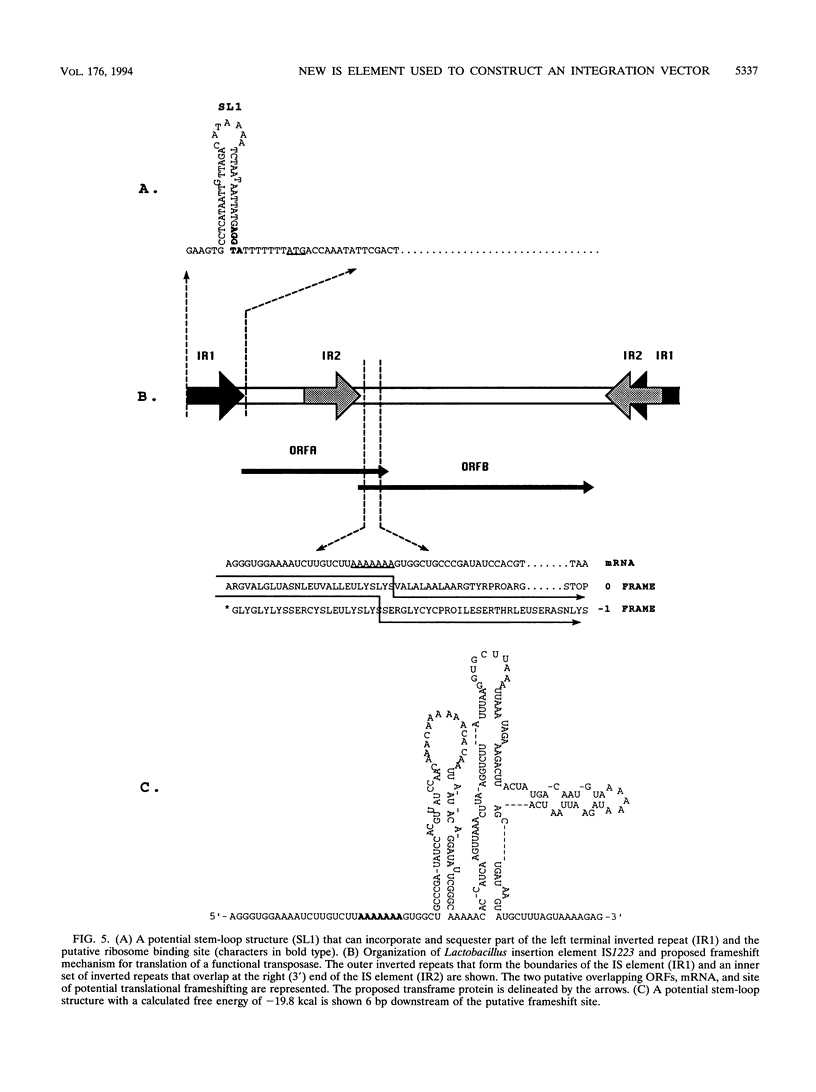
An insertion sequence (IS) element from Lactobacillus johnsonii was isolated, characterized, and exploited to construct an IS-based integration vector. L. johnsonii NCK61, a high-frequency conjugal donor of bacteriocin production (Laf+) and immunity (Lafr), was transformed to erythromycin resistance (Emr) with the shuttle vector pSA3. The NCK61 conjugative functions were used to mobilize pSA3 into a Laf- Lafs EMs recipient. DNA from the Emr transconjugants transformed into Escherichia coli MC1061 yielded a resolution plasmid with the same size as that of pSA3 with a 1.5-kb insertion. The gram-positive replication region of the resolution plasmid was removed to generate a pSA3-based suicide vector (pTRK327) bearing the 1.5-kb insert of Lactobacillus origin. Plasmid pTRK327 inserted randomly into the chromosomes of both Lactobacillus gasseri ATCC 33323 and VPI 11759. No homology was detected between plasmid and total host DNAs, suggesting a Rec-independent insertion. The DNA sequence of the 1.5-kb region revealed the characteristics of an IS element (designated IS1223): a length of 1,492 bp; flanking, 25-bp, imperfect inverted repeats; and two overlapping open reading frames (ORFs). Sequence comparisons revealed 71.1% similarity, including 35.7% identity, between the deduced ORFB protein of the E. coli IS element IS150 and the putative ORFB protein encoded by the Lactobacillus IS element. A putative frameshift site was detected between the overlapping ORFs of the Lactobacillus IS element. It is proposed that, similar to IS150, IS1223 produces an active transposase via translational frameshifting between two tandem, overlapping ORFs.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Atkins J. F., Weiss R. B., Gesteland R. F. Ribosome gymnastics--degree of difficulty 9.5, style 10.0. Cell. 1990 Aug 10;62(3):413–423. doi: 10.1016/0092-8674(90)90007-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhowmik T., Fernández L., Steele J. L. Gene replacement in Lactobacillus helveticus. J Bacteriol. 1993 Oct;175(19):6341–6344. doi: 10.1128/jb.175.19.6341-6344.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas I., Gruss A., Ehrlich S. D., Maguin E. High-efficiency gene inactivation and replacement system for gram-positive bacteria. J Bacteriol. 1993 Jun;175(11):3628–3635. doi: 10.1128/jb.175.11.3628-3635.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler M., Fayet O. Translational frameshifting in the control of transposition in bacteria. Mol Microbiol. 1993 Feb;7(4):497–503. doi: 10.1111/j.1365-2958.1993.tb01140.x. [DOI] [PubMed] [Google Scholar]
- Clark A. J., Warren G. J. Conjugal transmission of plasmids. Annu Rev Genet. 1979;13:99–125. doi: 10.1146/annurev.ge.13.120179.000531. [DOI] [PubMed] [Google Scholar]
- Dao M. L., Ferretti J. J. Streptococcus-Escherichia coli shuttle vector pSA3 and its use in the cloning of streptococcal genes. Appl Environ Microbiol. 1985 Jan;49(1):115–119. doi: 10.1128/aem.49.1.115-119.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis M. A., Simons R. W., Kleckner N. Tn10 protects itself at two levels from fortuitous activation by external promoters. Cell. 1985 Nov;43(1):379–387. doi: 10.1016/0092-8674(85)90043-1. [DOI] [PubMed] [Google Scholar]
- Derbyshire K. M., Grindley N. D. Binding of the IS903 transposase to its inverted repeat in vitro. EMBO J. 1992 Sep;11(9):3449–3455. doi: 10.1002/j.1460-2075.1992.tb05424.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodd I. B., Egan J. B. Improved detection of helix-turn-helix DNA-binding motifs in protein sequences. Nucleic Acids Res. 1990 Sep 11;18(17):5019–5026. doi: 10.1093/nar/18.17.5019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dower W. J., Miller J. F., Ragsdale C. W. High efficiency transformation of E. coli by high voltage electroporation. Nucleic Acids Res. 1988 Jul 11;16(13):6127–6145. doi: 10.1093/nar/16.13.6127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fayet O., Ramond P., Polard P., Prère M. F., Chandler M. Functional similarities between retroviruses and the IS3 family of bacterial insertion sequences? Mol Microbiol. 1990 Oct;4(10):1771–1777. doi: 10.1111/j.1365-2958.1990.tb00555.x. [DOI] [PubMed] [Google Scholar]
- Gay P., Le Coq D., Steinmetz M., Berkelman T., Kado C. I. Positive selection procedure for entrapment of insertion sequence elements in gram-negative bacteria. J Bacteriol. 1985 Nov;164(2):918–921. doi: 10.1128/jb.164.2.918-921.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guyer M. S. The gamma delta sequence of F is an insertion sequence. J Mol Biol. 1978 Dec 15;126(3):347–365. doi: 10.1016/0022-2836(78)90045-1. [DOI] [PubMed] [Google Scholar]
- Haandrikman A. J., van Leeuwen C., Kok J., Vos P., de Vos W. M., Venema G. Insertion elements on lactococcal proteinase plasmids. Appl Environ Microbiol. 1990 Jun;56(6):1890–1896. doi: 10.1128/aem.56.6.1890-1896.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holo H., Nes I. F. High-Frequency Transformation, by Electroporation, of Lactococcus lactis subsp. cremoris Grown with Glycine in Osmotically Stabilized Media. Appl Environ Microbiol. 1989 Dec;55(12):3119–3123. doi: 10.1128/aem.55.12.3119-3123.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebs M. P., Reznikoff W. S. Transcriptional and translational initiation sites of IS50. Control of transposase and inhibitor expression. J Mol Biol. 1986 Dec 20;192(4):781–791. doi: 10.1016/0022-2836(86)90028-8. [DOI] [PubMed] [Google Scholar]
- Kulkosky J., Jones K. S., Katz R. A., Mack J. P., Skalka A. M. Residues critical for retroviral integrative recombination in a region that is highly conserved among retroviral/retrotransposon integrases and bacterial insertion sequence transposases. Mol Cell Biol. 1992 May;12(5):2331–2338. doi: 10.1128/mcb.12.5.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Bourgeois P., Lautier M., Mata M., Ritzenthaler P. New tools for the physical and genetic mapping of Lactococcus strains. Gene. 1992 Feb 1;111(1):109–114. doi: 10.1016/0378-1119(92)90610-2. [DOI] [PubMed] [Google Scholar]
- Le Bourgeois P., Lautier M., Ritzenthaler P. Chromosome mapping in lactic acid bacteria. FEMS Microbiol Rev. 1993 Sep;12(1-3):109–123. doi: 10.1111/j.1574-6976.1993.tb00014.x. [DOI] [PubMed] [Google Scholar]
- Leer R. J., Christiaens H., Verstraete W., Peters L., Posno M., Pouwels P. H. Gene disruption in Lactobacillus plantarum strain 80 by site-specific recombination: isolation of a mutant strain deficient in conjugated bile salt hydrolase activity. Mol Gen Genet. 1993 May;239(1-2):269–272. doi: 10.1007/BF00281627. [DOI] [PubMed] [Google Scholar]
- Machida C., Machida Y. Regulation of IS1 transposition by the insA gene product. J Mol Biol. 1989 Aug 20;208(4):567–574. doi: 10.1016/0022-2836(89)90148-4. [DOI] [PubMed] [Google Scholar]
- Matsutani S., Ohtsubo H., Maeda Y., Ohtsubo E. Isolation and characterization of IS elements repeated in the bacterial chromosome. J Mol Biol. 1987 Aug 5;196(3):445–455. doi: 10.1016/0022-2836(87)90023-4. [DOI] [PubMed] [Google Scholar]
- Muriana P. M., Klaenhammer T. R. Conjugal Transfer of Plasmid-Encoded Determinants for Bacteriocin Production and Immunity in Lactobacillus acidophilus 88. Appl Environ Microbiol. 1987 Mar;53(3):553–560. doi: 10.1128/aem.53.3.553-560.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novel M., Huang D. C., Novel G. Transposition of the Streptococcus lactis ssp. lactis Z270 lactose plasmid to pVA797: demonstration of an insertion sequence and its relationship to an inverted repeat sequence isolated by self-annealing. Biochimie. 1988 Apr;70(4):543–551. doi: 10.1016/0300-9084(88)90091-0. [DOI] [PubMed] [Google Scholar]
- Ohtsubo H., Ohtsubo E. Isolation of inverted repeat sequences, including IS1, IS2, and IS3, in Escherichia coli plasmids. Proc Natl Acad Sci U S A. 1976 Jul;73(7):2316–2320. doi: 10.1073/pnas.73.7.2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polard P., Prère M. F., Chandler M., Fayet O. Programmed translational frameshifting and initiation at an AUU codon in gene expression of bacterial insertion sequence IS911. J Mol Biol. 1991 Dec 5;222(3):465–477. doi: 10.1016/0022-2836(91)90490-w. [DOI] [PubMed] [Google Scholar]
- Polzin K. M., McKay L. L. Development of a lactococcal integration vector by using IS981 and a temperature-sensitive lactococcal replication region. Appl Environ Microbiol. 1992 Feb;58(2):476–484. doi: 10.1128/aem.58.2.476-484.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polzin K. M., McKay L. L. Identification, DNA sequence, and distribution of IS981, a new, high-copy-number insertion sequence in lactococci. Appl Environ Microbiol. 1991 Mar;57(3):734–743. doi: 10.1128/aem.57.3.734-743.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polzin K. M., Romero D., Shimizu-Kadota M., Klaenhammer T. R., McKay L. L. Copy number and location of insertion sequences ISS1 and IS981 in lactococci and several other lactic acid bacteria. J Dairy Sci. 1993 May;76(5):1243–1252. doi: 10.3168/jds.S0022-0302(93)77453-6. [DOI] [PubMed] [Google Scholar]
- Polzin K. M., Shimizu-Kadota M. Identification of a new insertion element, similar to gram-negative IS26, on the lactose plasmid of Streptococcus lactis ML3. J Bacteriol. 1987 Dec;169(12):5481–5488. doi: 10.1128/jb.169.12.5481-5488.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prère M. F., Chandler M., Fayet O. Transposition in Shigella dysenteriae: isolation and analysis of IS911, a new member of the IS3 group of insertion sequences. J Bacteriol. 1990 Jul;172(7):4090–4099. doi: 10.1128/jb.172.7.4090-4099.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raya R. R., Fremaux C., De Antoni G. L., Klaenhammer T. R. Site-specific integration of the temperate bacteriophage phi adh into the Lactobacillus gasseri chromosome and molecular characterization of the phage (attP) and bacterial (attB) attachment sites. J Bacteriol. 1992 Sep;174(17):5584–5592. doi: 10.1128/jb.174.17.5584-5592.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero D. A., Klaenhammer T. R. Characterization of insertion sequence IS946, an Iso-ISS1 element, isolated from the conjugative lactococcal plasmid pTR2030. J Bacteriol. 1990 Aug;172(8):4151–4160. doi: 10.1128/jb.172.8.4151-4160.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero D. A., Klaenhammer T. R. IS946-mediated integration of heterologous DNA into the genome of Lactococcus lactis subsp. lactis. Appl Environ Microbiol. 1992 Feb;58(2):699–702. doi: 10.1128/aem.58.2.699-702.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheirlinck T., Mahillon J., Joos H., Dhaese P., Michiels F. Integration and expression of alpha-amylase and endoglucanase genes in the Lactobacillus plantarum chromosome. Appl Environ Microbiol. 1989 Sep;55(9):2130–2137. doi: 10.1128/aem.55.9.2130-2137.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz E., Herberger C., Rak B. Second-element turn-on of gene expression in an IS1 insertion mutant. Mol Gen Genet. 1988 Feb;211(2):282–289. doi: 10.1007/BF00330605. [DOI] [PubMed] [Google Scholar]
- Schäfer A., Jahns A., Geis A., Teuber M. Distribution of the IS elements ISS1 and IS904 in lactococci. FEMS Microbiol Lett. 1991 May 15;64(2-3):311–317. doi: 10.1111/j.1574-6968.1991.tb04681.x. [DOI] [PubMed] [Google Scholar]
- Scordilis G. E., Ree H., Lessie T. G. Identification of transposable elements which activate gene expression in Pseudomonas cepacia. J Bacteriol. 1987 Jan;169(1):8–13. doi: 10.1128/jb.169.1.8-13.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekine Y., Nagasawa H., Ohtsubo E. Identification of the site of translational frameshifting required for production of the transposase encoded by insertion sequence IS 1. Mol Gen Genet. 1992 Nov;235(2-3):317–324. doi: 10.1007/BF00279376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekine Y., Ohtsubo E. Frameshifting is required for production of the transposase encoded by insertion sequence 1. Proc Natl Acad Sci U S A. 1989 Jun;86(12):4609–4613. doi: 10.1073/pnas.86.12.4609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu-Kadota M., Flickinger J. L., Chassy B. M. Evidence that Lactobacillus casei insertion element ISL1 has a narrow host range. J Bacteriol. 1988 Oct;170(10):4976–4978. doi: 10.1128/jb.170.10.4976-4978.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu-Kadota M., Kiwaki M., Hirokawa H., Tsuchida N. ISL1: a new transposable element in Lactobacillus casei. Mol Gen Genet. 1985;200(2):193–198. doi: 10.1007/BF00425423. [DOI] [PubMed] [Google Scholar]
- Simon R., Hötte B., Klauke B., Kosier B. Isolation and characterization of insertion sequence elements from gram-negative bacteria by using new broad-host-range, positive selection vectors. J Bacteriol. 1991 Feb;173(4):1502–1508. doi: 10.1128/jb.173.4.1502-1508.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smigielski A. J. Characterization of a plasmid involved with cointegrate formation and lactose metabolism in Lactococcus lactis subsp. lactis OZS1. Arch Microbiol. 1990;154(6):560–565. doi: 10.1007/BF00248837. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Tanskanen E. I., Tulloch D. L., Hillier A. J., Davidson B. E. Pulsed-Field Gel Electrophoresis of SmaI Digests of Lactococcal Genomic DNA, a Novel Method of Strain Identification. Appl Environ Microbiol. 1990 Oct;56(10):3105–3111. doi: 10.1128/aem.56.10.3105-3111.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmerman K. P., Tu C. P. Complete sequence of IS3. Nucleic Acids Res. 1985 Mar 25;13(6):2127–2139. doi: 10.1093/nar/13.6.2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trieu-Cuot P., Courvalin P. Nucleotide sequence of the transposable element IS15. Gene. 1984 Oct;30(1-3):113–120. doi: 10.1016/0378-1119(84)90111-2. [DOI] [PubMed] [Google Scholar]
- Vögele K., Schwartz E., Welz C., Schiltz E., Rak B. High-level ribosomal frameshifting directs the synthesis of IS150 gene products. Nucleic Acids Res. 1991 Aug 25;19(16):4377–4385. doi: 10.1093/nar/19.16.4377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerbib D., Jakowec M., Prentki P., Galas D. J., Chandler M. Expression of proteins essential for IS1 transposition: specific binding of InsA to the ends of IS1. EMBO J. 1987 Oct;6(10):3163–3169. doi: 10.1002/j.1460-2075.1987.tb02627.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerbib D., Polard P., Escoubas J. M., Galas D., Chandler M. The regulatory role of the IS1-encoded InsA protein in transposition. Mol Microbiol. 1990 Mar;4(3):471–477. doi: 10.1111/j.1365-2958.1990.tb00613.x. [DOI] [PubMed] [Google Scholar]
- Zerbib D., Prentki P., Gamas P., Freund E., Galas D. J., Chandler M. Functional organization of the ends of IS1: specific binding site for an IS 1-encoded protein. Mol Microbiol. 1990 Sep;4(9):1477–1486. [PubMed] [Google Scholar]





