Abstract
The crystal structure of the six NH2-terminal zinc fingers of Xenopus laevis transcription factor IIIA (TFIIIA) bound with 31 bp of the 5S rRNA gene promoter has been determined at 3.1 Å resolution. Individual zinc fingers are positioned differently in the major groove and across the minor groove of DNA to span the entire length of the duplex. These results show how TFIIIA can recognize several separated DNA sequences by using fewer fingers than necessary for continuous winding in the major groove.
Keywords: 5S rRNA gene, Xenopus laevis, crystallography
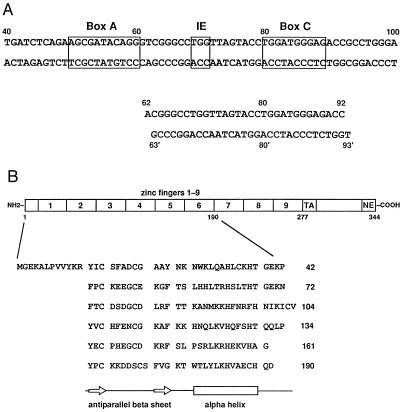
TFIIIA is an essential component of the RNA polymerase III (Pol III) transcription initiation complex for 5S rRNA in Xenopus laevis oocytes (1–3). TFIIIA also participates in the nuclear export (4) and storage of 5S rRNA, with which it forms a stable cytoplasmic 7S particle (5). The DNA-binding site for a single TFIIIA protein extends over 55 bp of the 5S rRNA gene promoter (6, 7). This site lies within the 5S rRNA coding sequence itself. It is effectively a tripartite promoter (8) containing separated “box A,” “intermediate element” (IE), and “box C” sequences (Fig. 1A). Similar regulatory elements exist in tRNA gene promoters. Mapping the details of this extensive protein–DNA interaction using chemical, biochemical, and genetic techniques has continued for almost 20 years (1–3, 9, 10). The discovery of nine zinc fingers in TFIIIA (11, 12) led to the notion of a transcription factor with repeated modules in its DNA-binding domain (Fig. 1B).
Figure 1.
Sequences of the DNA and protein used for crystallization. (A) Pol III elements within the X. laevis oocyte 5S rRNA ICR (base pairs +43 to +97) are shown boxed. The 31-bp duplex is numbered according to the 5S rRNA gene. (B) The six-finger protein corresponds to amino acid residues 1–190 of X. laevis TFIIIA (42, 43). Zinc fingers are aligned to show their secondary structure. Beta sheet is indicated by open arrows and the alpha helix is indicated as an open box. The “TA” region of TFIIIA is required for transcription activation (56) and “NE” is required for nuclear export (4).
Our present knowledge of how zinc fingers bind specifically to DNA comes largely from several x-ray structures (13–18). In all of these protein–DNA complexes, there are contiguous zinc finger interactions with base pairs in the major groove. In Zif268, for example, three fingers recognize successive, overlapping base pair quartets in the major groove, covering a total of 10 bp. In the DNA complex of a five-finger segment from Gli, the first finger lies outside the major groove and makes no DNA contacts. The remaining fingers wrap in the major groove rather like those of Zif268. An extension of this mode of binding is not sufficient to explain the size of the TFIIIA-binding site, however (see, for example, models proposed in refs. 9 and 10). The NH2-terminal six zinc fingers of TFIIIA bound in a complex with 31 bp of the 5S rRNA gene has been reconstituted (19) and crystallized. We report here the structure of the complex at 3.1 Å resolution (Table 1) and describe how its zinc fingers interact with DNA in different ways. An NMR structure of fingers 1–2–3 bound to DNA has recently been published (20, 21).
Table 1.
Statistics from the crystallographic analysis
| Native | Derivative 1*
|
Derivative 2*
|
||||
|---|---|---|---|---|---|---|
| Data set 1 | Data set 2 | Data set 3 | Data set 4 | Data set 5 | ||
| Data collection | ||||||
| Observed reflections | 126,746 | 74,611 | 86,629 | 47,898 | 33,420 | 56,366 |
| Unique reflections | 19,034 | 20,631 | 21,289 | 22,676 | 17,871 | 22,863 |
| Completeness, % | 77.4† | 96.0 | 97.6 | 92.8 | 70.6 | 95.6 |
| Rmerge, % | 12.8 | 15.2 | 11.0 | 12.0 | 15.5 | 13.1 |
| Mean I/σ | 7.0 | 5.1 | 8.4 | 6.0 | 4.0 | 5.0 |
| MIR analysis | ||||||
| Resolution, Å | 40–3.1 | |||||
| Mean isomorphous difference, % | 14.7 | 12.6 | 20.0 | 21.8 | 18.6 | |
| Phasing power | 1.48 | 1.57 | 1.61 | 1.79 | 1.93 | |
| Cullis R-factor | 0.82 | 0.81 | 0.82 | 0.75 | 0.74 | |
| Mean overall figure of merit | 0.47 | |||||
| Refinement | ||||||
| Resolution, Å | 8–3.1 | |||||
| R-factor, % | 29.7 | |||||
| Free R-factor, % | 33.7 | |||||
| Reflections with |F| > 2σ | 17,027 | |||||
| Nonhydrogen atoms | 5,474 | |||||
| Bond lengths RMS deviation, Å | 0.02 | |||||
| Bond angles RMS deviation, degrees | 2.35 | |||||
Positions of 5IdU substitution in derivative 1 are T73, T76, and T93′, and in derivative 2 are T73, T76, and T88′.
After local filtering with an Ihkl/σhkl cutoff of 1.0 to eliminate regions of unreliable data. Crystallographic R-factor = ∑ |Fobs − Fcalc|/∑|Fobs|, calculated using anisotropically sharpened data.
MATERIALS AND METHODS
Protein and DNA Oligonucleotides.
Recombinant TFIIIA (amino acid residues 1–190) was produced from plasmid pRSET B (Invitrogen) in Escherichia coli BL21(DE3). After sonication the protein was extracted in 7 M urea from cell pellets and purified on Bio-Rex 70 (Bio-Rad) and heparin Sepharose columns. Synthetic oligonucleotides were purified by MonoQ (Pharmacia) chromatography in 7 M urea. Thymines were replaced by 5-iododeoxyuracil (5IdU) at specific positions: T73 and T76 (noncoding strand), and T88′ or T93′ (coding strand), for two triple heavy atom derivatives.
Reconstitution and Crystallization.
The protein–DNA complex was reconstituted by stepwise dilution from 0.75–0.25 M NaCl at 25°C (19). Crystals grew in hanging drops on silanized plastic coverslips from 165 mM NaCl, 35 mM sodium acetate, 3.2 mM DTT, 9.2% (vol/vol) glycerol, 1.8 mM NaN3, 1.8 mM cadaverine-2 HCl, 5.5 mM Tris⋅HCl, pH 8.0, and 22.5% PEG 4000 at 18°C. The complex crystallized in the space group P1 with unit cell parameters a = 64.2 Å, b = 64.7 Å, c = 78.0 Å, α = 90.1°, β = 93.0°, γ = 103.0°. Two complexes are present in the unit cell, which contains 72% solvent.
Cryocrystallography.
Invariably doubled crystals were split under polarized light and cryoprotectant was introduced in steps over 48 hr to reach the final conditions, which were mother liquor supplemented with 215 mM NaCl, 10% sucrose, and 15% (vol/vol) glycerol. Crystals (25- to 50-micron thickness) were frozen in nylon loops (10–0 Ethilon suture) by plunging into liquid nitrogen. Data were collected at −160°C with a MAR image-plate detector (MAR Research, Hamburg) at a wavelength of 1.283 Å (the K-edge of zinc) at the National Synchrotron Light Source beamline X12B at Brookhaven. Ice rings were deleted from the MAR images, and intensities were integrated and merged with denzo/scalepack programs (22).
Image-Seeking Analysis.
The structure was determined at low resolution by multiple isomorphous replacement (MIR), by using two derivatives (Table 1). A difference Patterson synthesis for derivative 1, calculated with the FFT program (23), was searched with a model of the three iodine atoms derived from B-form DNA. The correct constellation of six Patterson peaks has noncrystallographic symmetry—a twofold rotation around and a translation along a direction parallel to the b axis. Thirty-eight thousand potential solutions were evaluated with vecref (23), and MIR phases were calculated by mlphare (23). Two data sets from one derivative and three from the other derivative were used in the final phasing (Table 1).
Anomalous Difference Fourier.
Zinc anomalous scattering data were derived from a merged data set of four native crystals. Anomalous differences |F+ − F−|hkl > 30% of |F+ + F−|hkl/2 were rejected before calculating Fourier maps with MIR phases.
Refinement and Model Building.
Phases were extended from 6 to 4.5 Å by positioning base pairs into an MIR map averaged with rave (24). Homologous zinc fingers taken from known crystal structures (13–18) were positioned into appropriate electron density and refined by using the real-space, rigid-body procedure in the o graphics program (25). Combined phases from MIR and the partial model were generated by sigmaa (23) as the starting point for several cycles of mask refinement, averaging, solvent flattening, and rigid-body refinement. Phases were extended to 3.1 Å resolution by an iterative procedure involving (i) twofold averaging, solvent flattening, and histogram matching with the dm program (26); (ii) model rebuilding with program o, using a custom zinc finger lego library, and (iii) positional refinement into anisotropically scaled, B-factor sharpened data with x-plor (27). At late stages in model rebuilding, “omit” maps were generated by deleting individual fingers from the model.
RESULTS
Description of the Structure.
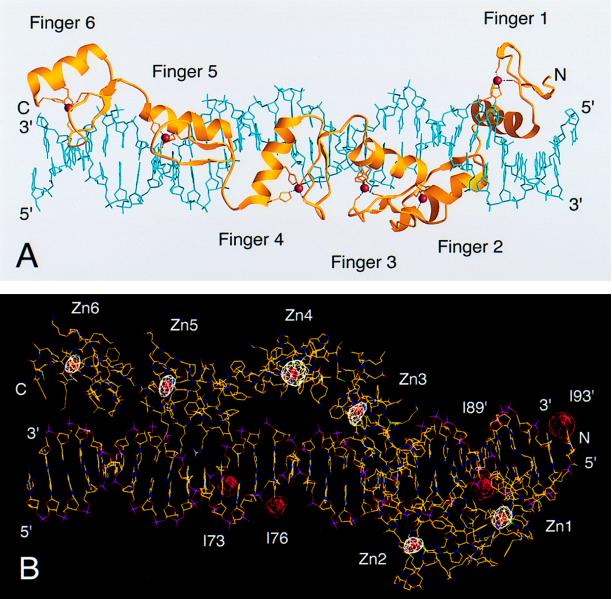
The crystal structure shows that the six-finger protein stretches along the entire length of the 31-bp duplex. The current protein model includes amino acids 10–188 of TFIIIA (Fig. 1B). Residues 1–9, 161, and 189–190 are disordered in the crystal. In the complex, fingers 1–2–3 adopt a completely different configuration than do fingers 4–5–6 (Fig. 2A). Fingers 1–2–3, which are separated by typical linker sequences, wrap smoothly around the major groove of DNA rather like those of Zif268 (13, 16). Contacts are made with DNA bases mainly on the noncoding strand of the 5S rRNA gene. In contrast, fingers 4–5–6, which run along one side of the DNA double helix, form an open, extended structure. Of these, only finger 5 makes contacts with bases in the major groove. The two flanking fingers, 4 and 6, straddle the neighboring minor grooves and appear to serve primarily as spacer elements in DNA recognition. In this way the six-finger protein binds in a precise manner to the separated IE and box C sequences (8).
Figure 2.
Structure of the six-finger TFIIIA–DNA complex. (A) A ribbons (57) representation, in which alpha helices and beta sheets of TFIIIA are colored yellow; Zn(II) ions are red spheres; and the DNA double helix is light blue. (B) Crystallographic assignment of zinc fingers and DNA bases to locations within the complex. A zinc anomalous difference Fourier map (calculated at 4 Å resolution, white contour levels >4σ) and 5IdU difference Fourier maps (calculated at 5 Å resolution, red contour levels >5σ) are shown superimposed on a molecular model generated with the program o (25). Carbon atoms are colored yellow; oxygen, red; nitrogen, blue; sulfur, green; and phosphate groups, magenta. The direction of view in B is oriented approximately perpendicular to the direction in A.
The DNA is essentially B-form with a mean helical twist of 34.3° and a rise per base pair of 3.33 Å. Its sequence corresponds to base pairs +63 to +92 of the internal control region (ICR) of the 5S rRNA gene. Terminal 5′-overhanging bases are involved in normal Watson–Crick base pairs with neighboring duplexes so as to form continuous columns of DNA in the crystal lattice. Analysis of the double helix with the program curves (28) shows that there are three localized bends (29) of 16.7°, 24.4°, and 18.4° at base pairs +70, +85, and +90. Fingers 5, 2, and 1 interact with these positions, respectively. Furthermore, as a result of zinc finger binding there are increases in the depth and width of the major groove (30).
Each TFIIIA finger is folded in the classical way (31, 32) around a Zn(II) ion, including finger 6, which lacks some of the conserved amino acid residues. The positions of the six Zn(II) ions were determined independently of the protein structure from an anomalous difference Fourier synthesis. These metal sites are also present in the electron density and indicate the correct path and fold of the polypeptide chain (Fig. 2B).
The consensus pentapeptide linker sequence Thr-Gly-Glu-Lys-Pro, frequently associated with major-groove binding fingers, appears only twice in this NH2-terminal segment of TFIIIA. As expected, these linkers, 1–2 and 2–3, do indeed connect fingers that interact with bases in the major groove. The remaining linkers, 3–4, 4–5, and 5–6, have different structures and sequences, which permit the extended configuration for fingers 4, 5, and 6. Linkers 3–4 and 4–5 fold in ways that bring several hydrophobic amino acids into proximity. Five residues of linker 3–4—Ile-100, Ile-102, Cys-103, Val-104, and Val-106—form a hydrophobic cluster. Likewise four residues at the interface of fingers 4–5—Val-124, Phe-127, Pro-134, and Tyr-135—come together in another hydrophobic cluster, and Phe-127 also makes van der Waals contacts with Pro-134.
Zinc Finger–DNA Interactions.
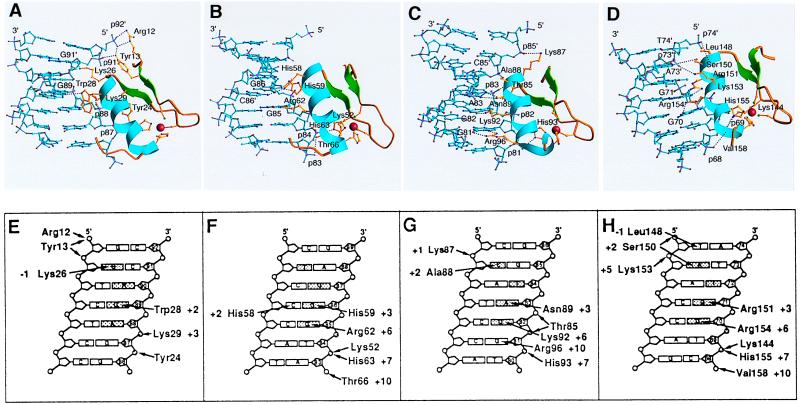
For the most part, the alpha helices of fingers 2, 3, and 5 interact with DNA as in previously analyzed structures, with side chains contacting at least two of four consecutive base pairs in the major groove (Fig. 3). Base pair quartets that interact with adjacent fingers overlap by one, and backbone contacts from successive fingers overlap even more extensively. In the previously analyzed complexes, most of the major-groove contacts occur between three bases on one strand and amino acid side chains at alpha helix positions +6, +3, and −1, and the opposite-strand base of the fourth base pair in the quartet may contact the side chain of alpha helix position +2 (see shaded bases in Fig. 3 E–H). In our structure, the “canonical” +6 and +3 contacts are made by fingers 2, 3, and 5, which also have a commonly found His (+7)–phosphate interaction. The +2 and −1 contacts are less standard, and in finger 3, the site is extended by an arginine–guanine interaction from the +10 position of the alpha helix (Fig. 3 C and G).
Figure 3.
DNA major-groove contacts with each of the zinc fingers 1, 2, 3, and 5. (A–D) The zinc fingers are placed in similar orientations. The protein is shown as a ribbon with alpha helix, blue, and beta sheet, green. The DNA is light blue. The amino acid side chains that contact nucleotide bases are yellow, and hydrogen-bond contacts are shown as dotted lines. Oxygen atoms are red, and nitrogen, magenta. (E–H) The major groove of DNA is represented schematically in cylindrical projection. The noncoding strand is numbered as in the 5S rRNA gene. Nucleotide bases of the “canonical” quartet for contacts by zinc fingers in previously analyzed structures are shown shaded, as are two phosphates that frequently receive hydrogen bonds. Contacts between amino acids and DNA are drawn as arrows.
Finger 1 is oriented similarly to fingers 2, 3, and 5 with respect to the groove, but it is displaced by more than 4 Å toward the COOH terminus of its recognition helix (Fig. 3 A and E). As a result, the indole Nɛ of Trp-28 at position +2 in the helix lies opposite the O6 of G89, and Lys-29 (+3), which would normally contact this guanine, forms a salt bridge with phosphate-88 instead. Lys-26 (−1) interacts with the opposite-strand guanine “vacated” by the +2 tryptophan. The shifted position of finger 1 requires the small Ala-32 side chain at position +6 to avoid steric interference, and it places Tyr-24 rather than His-33 within hydrogen-bond distance of phosphate-87.
Together, fingers 1–2–3 bind within an 11-bp region located between positions +81 and +91 of the ICR, specifying the DNA sequence GGANGGNNGNN (noncoding strand) and NNNNCCNNNNG (coding strand). The structure agrees well with the earlier identification of the Pol III promoter element box C that was derived from site-directed mutagenesis of the 5S rRNA gene (8). The local details of finger conformation and DNA contacts, seen in a recent NMR structure of fingers 1–2–3 bound to 15 bp of DNA (20, 21), closely match our x-ray structure of the longer fragment. The relative orientation and position of finger 1 with respect to fingers 2 and 3 are somewhat different, however. In the x-ray structure, fingers 1 and 2 are not in direct contact, whereas in the NMR structure, they close down against each other, with a concomitant difference in the DNA conformation so that base pair contacts are unperturbed.
Finger 5 binds to bases in the major groove at the IE element, positions +70 to +73, 7 bp upstream of box C. A standard major-groove finger interaction is supplemented by contacts between Leu-148 (−1) and the base of T74′, Ser-150 (+2) and phosphate-74′, and Lys-153 and phosphate-73′. The DNA sequence specified by finger 5 is GGNNN (noncoding strand) and NNNAT (coding strand), the consensus IE sequence (8).
From a structural standpoint there is nothing different about fingers 4 and 6. They do not wrap around the double helix but instead traverse the minor groove. As spacers they increase the range of the TFIIIA protein, making possible a more economical use of fingers in binding to the separate promoter elements, IE and box C. In spanning the minor groove a few contacts are made with the DNA backbone. Gln-121 and conserved Tyr-135 of finger 4 both contact phosphate-75′, and Lys-175 of finger 6 contacts phosphate-68.
The DNA contacts present in our x-ray structure are consistent with previous biochemical and genetic analyses. A number of guanine bases in the TFIIIA–DNA complex are protected from methylation (33). Prior methylation of certain guanine bases or ethylation of several phosphates interferes with the binding of TFIIIA (10, 34). Site-directed mutagenesis of base pairs within the ICR decreases transcriptional activity (8, 35) or lowers TFIIIA binding (36, 37). Some amino acid mutations in TFIIIA linkers (38–40) or zinc fingers (41) decrease TFIIIA binding to the ICR. A recent summary of biochemical and genetic studies on fingers 1–2–3 may be found in ref. 21.
DISCUSSION
TFIIIA specificity is highly conserved. This conservation is apparent at positions in the recognition helix that are involved in binding to DNA (Table 2) in eight aligned TFIIIA sequences (42–48). Similarly, the nucleotide bases that make contacts with fingers 1–2–3 and 5 are invariant with some exceptions in Ictalurus puntatus and Saccharomyces cerevisiae 5S rRNA genes (45, 49–53). Finger 1 has almost no amino acid sequence variation. Substitutions are often conservative. Moreover, to the extent that Lys replaced by Arg may still specify guanine and that Asn replaced by Gln may specify adenine, the substitutions do not affect DNA recognition. In S. cerevisiae TFIIIA the sequence identity is limited to the recognition helix of finger 2. Nevertheless, the same methylation pattern of guanine residues in the ICR that interfere with binding of fingers 1–2–3 was also found (54).
Table 2.
Comparison of DNA-recognition helices and linkers in TFIIIA sequences
| TFIIIA sequence | Amino acid alignment | |||
|---|---|---|---|---|
| Helix residues −1, +2, +3, and +6 in fingers: 1, 2, 3, and 5* | ||||
| Xenopus laevis | KWKA | SHHR | TANK | LSRR |
| Xenopus borealis | KWKA | SHHR | TANK | VSCR |
| Rana pipiens | KWKA | TFHR | TTNK | SSRR |
| Rana catesbeiana | KWKA | TFHR | TTNK | SSRR |
| Bufo americanus | KRKA | THHR | TSNL | SSRR |
| Homo sapiens | KWKA | RYHR | TSNK | SSKR |
| Ictalurus punctatus | KWKA | TCQR | SAGN | TKKK |
| Saccharomyces cerevisiae | RISE | KSHR | TQRR | RYRN |
| Linker sequences: 1–2, 2–3, 3–4, 4–5, and 5–6† | ||||
| Xenopus laevis | TGEKP | TGEKN | NIKICV | TQQLP | AG |
| Xenopus borealis | TGEKP | TGEKN | NLQLCV | TQQLO | AG |
| Rana pipiens | TGERP | TGEKP | LSPSLI | TNQQP | AG |
| Rana catesbeiana | TGERP | TGEKP | LSPSLI | TNQQP | AG |
| Bufo americanus | TGERP | TGEKP | SSPAQI | TNQQP | AG |
| Homo sapiens | TGERP | TGEKP | QNQQKQ | TNEPL | EG |
| Ictalurus punctatus | TGLRP | SGKKP | QHKEKH | MNQLP | DV |
| Saccharomyces cerevisiae | QGLRA | SDTKP | TKS | LHK | HDPEVENP |
The characteristics of individual linkers are the same for various TFIIIA sequences (see Table 2). This conserved pattern points to a common structural organization for these proteins. In addition, invariant helix residues and corresponding DNA bases suggest that the topology of fingers 1–6 in other TFIIIA–DNA complexes will be identical to our structure.
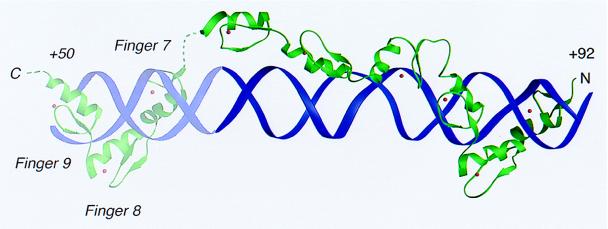
Fingers 7–8–9, not present in our structure, bind to the Pol III element box A. It has been proposed, based on results from DNA methylation protection and binding interference and on site-directed mutagenesis experiments (8, 33–37), that these fingers wrap, like fingers 1–2–3, around the major groove of base pairs +48 to +62 (9, 10). In the model shown in Fig. 4, fingers 7–8–9 have been placed so that Arg-271 at helix position +6 in finger 9 can recognize G51. Finger 6 can then connect to finger 7 with only a small displacement from its position in our crystal structure. We note that linkers 7–8 and 8–9 have amino acid sequences resembling the Thr-Gly-Glu-Lys-Pro consensus characteristic of sets of fingers that wrap around the major groove.
Figure 4.
A model for the nine-finger TFIIIA–DNA complex. The DNA double helix (purple) and the TFIIIA zinc fingers (green) are shown as ribbons. Zn(II) ions are red spheres. The positioning of the COOH-terminal fingers 7–8–9 in the major groove is derived from biochemical analysis of the ICR (9, 10).
The x-ray structure of the TFIIIA–DNA complex shows how zinc fingers have been deployed to bind to separated promoter elements. Local folding of the protein orients fingers with respect to each other for a “custom fit” to the extended site. In this sort of design, some fingers will contact base pairs and some will not. It is likely that other multifingered proteins will use a similar strategy to recognize regulatory elements in DNA. Bridging fingers may also serve additional functions in the multiprotein assemblies that activate transcription. Fingers 4–5–6 in this structure form a continuous, platform-like surface, which could dock against other components of a Pol III transcription complex.
In our structure three of the four fingers insert in the major groove essentially in the manner previously seen in other complexes, but the fourth (finger 1) is displaced by about one base pair and has idiosyncratic interactions. Recent efforts to design zinc finger proteins with desired DNA specificity have concentrated on the recognition helix (55). Our structure further justifies the focus on a roughly standard orientation for this helix, but it also provides an alternative framework for the design of proteins that recognize an extended site. Mutagenesis and selection of linkers are likely to be particularly important in engineering such proteins.
Acknowledgments
We thank K. Clemens for providing a cDNA clone of TFIIIA (amino acid residues 1–190) and D. Setzer and J. Gottesfeld for sharing their experience with protein purification. We are indebted to R. Lawrence, R. Liddington, M. Rould, D. Rodgers, J.-H. Wang, and R. Marmorstein for advice on crystallographic computing and to M. Capel for help with data collection. R.T.N. was supported by the U.S. Army Medical Research and Material Command under DAMD17-94-J-4420. S.C.H. is an investigator in the Howard Hughes Medical Institute.
ABBREVIATIONS
- TFIIIA
transcription factor IIIA
- Pol III
RNA polymerase III
- IE
intermediate element
- ICR
internal control region
Footnotes
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, Chemistry Department, Brookhaven National Laboratory, Upton, NY 11973 (reference 1TF6).
References
- 1.Hanas J S, Gaskins C J, Smith J F, Ogilvie M K. Prog Nucleic Acid Res Mol Biol. 1992;43:205–239. doi: 10.1016/s0079-6603(08)61048-x. [DOI] [PubMed] [Google Scholar]
- 2.Pieler T, Theunissen O. Trends Biochem Sci. 1993;18:226–230. doi: 10.1016/0968-0004(93)90194-r. [DOI] [PubMed] [Google Scholar]
- 3.Shastry B S. J Cell Sci. 1996;109:535–539. doi: 10.1242/jcs.109.3.535. [DOI] [PubMed] [Google Scholar]
- 4.Friedell R A, Fischer V, Luhrmann R, Meyer B E, Meinkoth J L, Malius M H, Cullen B R. Proc Natl Acad Sci USA. 1996;93:2936–2940. doi: 10.1073/pnas.93.7.2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Picard B, Wegnez M. Proc Natl Acad Sci USA. 1979;76:241–245. doi: 10.1073/pnas.76.1.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Engelke D R, Ng S-Y, Shastry B S, Roeder R G. Cell. 1980;19:717–728. doi: 10.1016/s0092-8674(80)80048-1. [DOI] [PubMed] [Google Scholar]
- 7.Pelham H R B, Brown D D. Proc Natl Acad Sci USA. 1980;77:4170–4174. doi: 10.1073/pnas.77.7.4170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pieler T, Oei S-L, Hamm J, Engelke U, Erdmann V A. EMBO J. 1985;4:3751–3756. doi: 10.1002/j.1460-2075.1985.tb04144.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hayes J J, Tullius T D. J Mol Biol. 1992;227:407–417. doi: 10.1016/0022-2836(92)90897-s. [DOI] [PubMed] [Google Scholar]
- 10.Clemens K R, Liao X, Wolf V, Wright P E, Gottesfeld J M. Proc Natl Acad Sci USA. 1992;89:10822–10826. doi: 10.1073/pnas.89.22.10822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brown R S, Sander C, Argos P. FEBS Lett. 1985;186:271–274. doi: 10.1016/0014-5793(85)80723-7. [DOI] [PubMed] [Google Scholar]
- 12.Miller J, McLachlan A D, Klug A. EMBO J. 1985;4:1609–1614. doi: 10.1002/j.1460-2075.1985.tb03825.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pavletich N P, Pabo C O. Science. 1991;252:809–817. doi: 10.1126/science.2028256. [DOI] [PubMed] [Google Scholar]
- 14.Pavletich N P, Pabo C O. Science. 1993;261:1701–1707. doi: 10.1126/science.8378770. [DOI] [PubMed] [Google Scholar]
- 15.Fairall L, Schwabe J W R, Chapman L, Finch J T, Rhodes D. Nature (London) 1993;366:483–487. doi: 10.1038/366483a0. [DOI] [PubMed] [Google Scholar]
- 16.Elrod-Erickson M, Rould A M, Nekludova L, Pabo C O. Structure. 1996;4:1171–1180. doi: 10.1016/s0969-2126(96)00125-6. [DOI] [PubMed] [Google Scholar]
- 17.Houbaviy H B, Usheva A, Shenk T, Burley S K. Proc Natl Acad Sci USA. 1996;93:13577–13582. doi: 10.1073/pnas.93.24.13577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim C A, Berg J M. Nat Struct Biol. 1996;3:940–945. doi: 10.1038/nsb1196-940. [DOI] [PubMed] [Google Scholar]
- 19.Conlin R M, Brown R S. Methods Mol Biol. 1994;30:357–370. doi: 10.1385/0-89603-256-6:357. [DOI] [PubMed] [Google Scholar]
- 20.Foster M P, Wuttke D S, Radhakrishnan I, Case D A, Gottesfeld J M, Wright P E. Nat Struct Biol. 1997;4:605–608. doi: 10.1038/nsb0897-605. [DOI] [PubMed] [Google Scholar]
- 21.Wuttke D S, Foster M P, Case D A, Gottesfeld J M, Wright P E. J Mol Biol. 1997;273:183–206. doi: 10.1006/jmbi.1997.1291. [DOI] [PubMed] [Google Scholar]
- 22.Otwinowski Z, Minor W. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 23.Collaborative Computational Project, Number 4. Acta Crystallogr. 1994;D50:760–763. [Google Scholar]
- 24.Kleywegt G J, Jones T A. Proceedings of the CCP 4 Study Weekend. Daresbury, U.K.: SERC Daresbury; 1994. pp. 59–66. [Google Scholar]
- 25.Jones T A, Kjeldgaard K. o Computer Graphics Program. Sweden: Uppsala Univ.; 1993. [Google Scholar]
- 26.Cowtan K D. Joint CCP 4 ESF-EACBM Newsletter Protein Crystallogr. 1994;31:34–38. [Google Scholar]
- 27.Brünger A T. x-plor Version 3.8, A System for X-Ray Crystallography and NMR. New Haven, CT: Yale Univ. Press; 1996. [Google Scholar]
- 28.Lavery R, Sklenar H. J Biomol Struct Dyn. 1989;6:655–667. doi: 10.1080/07391102.1989.10507728. [DOI] [PubMed] [Google Scholar]
- 29.Zwieb C, Brown R S. Nucleic Acids Res. 1990;18:583–587. doi: 10.1093/nar/18.3.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nekludova L, Pabo C O. Proc Natl Acad Sci USA. 1994;91:6948–6952. doi: 10.1073/pnas.91.15.6948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Berg J. Proc Natl Acad Sci USA. 1988;85:99–102. doi: 10.1073/pnas.85.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee M S, Gippert G P, Soman K V, Case D A, Wright P E. Science. 1989;245:635–637. doi: 10.1126/science.2503871. [DOI] [PubMed] [Google Scholar]
- 33.Fairall L, Rhodes D, Klug A. J Mol Biol. 1986;192:577–591. doi: 10.1016/0022-2836(86)90278-0. [DOI] [PubMed] [Google Scholar]
- 34.Sakonju S, Brown D D. Cell. 1982;31:395–405. doi: 10.1016/0092-8674(82)90133-7. [DOI] [PubMed] [Google Scholar]
- 35.McConkey G A, Bogenhagen D F. Mol Cell Biol. 1987;7:486–494. doi: 10.1128/mcb.7.1.486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Veldhoen N, You Q M, Setzer D R, Romaniuk P J. Biochemistry. 1994;33:7568–7575. doi: 10.1021/bi00190a009. [DOI] [PubMed] [Google Scholar]
- 37.Rawlings S L, Matt G D, Huber P W. J Biol Chem. 1996;271:868–877. doi: 10.1074/jbc.271.2.869. [DOI] [PubMed] [Google Scholar]
- 38.Smith J F, Hawkins J, Leonard R E, Hanas J S. Nucleic Acids Res. 1991;19:6871–6876. doi: 10.1093/nar/19.24.6871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Choo Y, Klug A. Nucleic Acids Res. 1993;21:3341–3346. doi: 10.1093/nar/21.15.3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Clemens K R, Zhang P, Liao X, McBryant S J, Wright P E, Gottesfeld J M. J Mol Biol. 1994;244:23–35. doi: 10.1006/jmbi.1994.1701. [DOI] [PubMed] [Google Scholar]
- 41.Zang W Q, Veldhoen N, Romaniuk P J. Biochemistry. 1995;34:15545–15552. doi: 10.1021/bi00047a021. [DOI] [PubMed] [Google Scholar]
- 42.Ginsberg A M, King B O, Roeder R G. Cell. 1984;39:479–489. doi: 10.1016/0092-8674(84)90455-0. [DOI] [PubMed] [Google Scholar]
- 43.Taylor W, Jackson I J, Siegel N, Kumar A, Brown D D. Nucleic Acids Res. 1986;14:6185–6195. doi: 10.1093/nar/14.15.6185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gaskins C J, Hanas J S. Nucleic Acids Res. 1990;18:2117–2123. doi: 10.1093/nar/18.8.2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gaskins C J, Smith J F, Ogilvie M K, Hanas J S. Gene. 1992;120:197–206. doi: 10.1016/0378-1119(92)90094-6. [DOI] [PubMed] [Google Scholar]
- 46.Archambault J, Milne C A, Shappert K T, Baum B, Friesen J D, Segall J. J Biol Chem. 1992;267:3282–3288. [PubMed] [Google Scholar]
- 47.Arakawa H, Nagase H, Hayashi N, Ogawa M, Nagata M, Fujiwara T, Takahashi E, Shin S, Nakamura Y. Cytogenet Cell Genet. 1995;70:235–238. doi: 10.1159/000134041. [DOI] [PubMed] [Google Scholar]
- 48.Ogilvie M K, Hanas J S. Gene. 1997;203:103–112. doi: 10.1016/s0378-1119(97)00499-x. [DOI] [PubMed] [Google Scholar]
- 49.Wegnez M, Monier R, Denis H. FEBS Lett. 1972;25:13–20. doi: 10.1016/0014-5793(72)80443-5. [DOI] [PubMed] [Google Scholar]
- 50.Korn L J, Brown D D. Cell. 1978;15:1145–1156. doi: 10.1016/0092-8674(78)90042-9. [DOI] [PubMed] [Google Scholar]
- 51.Forget B G, Weissman S M. J Biol Chem. 1969;244:3148–3165. [PubMed] [Google Scholar]
- 52.Valenzuela P, Bell G I, Masiarz F R, DeGennaro, Rutter W J. Nature (London) 1977;267:641–643. doi: 10.1038/267641a0. [DOI] [PubMed] [Google Scholar]
- 53.Maxwell E S, Martin T E. Nucleic Acids Res. 1986;14:5741–5760. doi: 10.1093/nar/14.14.5741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rowland O, Segall J. J Biol Chem. 1996;271:12103–12110. doi: 10.1074/jbc.271.20.12103. [DOI] [PubMed] [Google Scholar]
- 55.Greisman H A, Pabo C O. Science. 1997;275:657–661. doi: 10.1126/science.275.5300.657. [DOI] [PubMed] [Google Scholar]
- 56.Mao X, Darby M K. Mol Cell Biol. 1993;13:7496–7506. doi: 10.1128/mcb.13.12.7496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Carson M. Methods Enzymol. 1997;277:493–505. [PubMed] [Google Scholar]