Abstract
Overexpression of the Tn5 transposase (Tnp) was found to be lethal to Escherichia coli. This killing was not caused by transposition or dependent on the transpositional or DNA binding competence of Tnp. Instead, it was strictly correlated with the presence of a wild-type N terminus. Deletions removing just two N-terminal amino acids of Tnp resulted in partial suppression of this effect, and deletions of Tnp removing 3 or 11 N-terminal amino acids abolished the killing effect. This cytotoxic effect of Tnp overexpression is accompanied by extensive filament formation (i.e., a defect in cell division) and aberrant nucleoid segregation. Four E. coli mutants were isolated which allow survival upon Tnp overexpression, and the mutations are located at four discrete loci. These suppressor mutations map near essential genes involved in cell division and DNA segregation. One of these mutations maps to a 4.5-kb HindIII region containing the ftsYEX (cell division) locus at 76 min. A simple proposition which accounts for all of these observations is that Tnp interacts with an essential E. coli factor affecting cell division and/or chromosome segregation and that overexpression of Tnp titrates this factor below a level required for viability of the cell. Furthermore, the N terminus of Tnp is necessary for this interaction. The possible significance of this phenomenon for the transposition process is discussed.
Full text
PDF









Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baker T. A., Mizuuchi K. DNA-promoted assembly of the active tetramer of the Mu transposase. Genes Dev. 1992 Nov;6(11):2221–2232. doi: 10.1101/gad.6.11.2221. [DOI] [PubMed] [Google Scholar]
- Boeckh C., Bade E. G., Delius H., Reeve J. N. Inhibition of bacterial segregation by early functions of phage mu and association of replication protein B with the inner cell membrane. Mol Gen Genet. 1986 Mar;202(3):461–466. doi: 10.1007/BF00333277. [DOI] [PubMed] [Google Scholar]
- Bukau B., Walker G. C. Delta dnaK52 mutants of Escherichia coli have defects in chromosome segregation and plasmid maintenance at normal growth temperatures. J Bacteriol. 1989 Nov;171(11):6030–6038. doi: 10.1128/jb.171.11.6030-6038.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLong A., Syvanen M. Membrane association of the Tnp and Inh proteins of IS50R. J Bacteriol. 1990 Sep;172(9):5516–5519. doi: 10.1128/jb.172.9.5516-5519.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLong A., Syvanen M. Trans-acting transposase mutant from Tn5. Proc Natl Acad Sci U S A. 1991 Jul 15;88(14):6072–6076. doi: 10.1073/pnas.88.14.6072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donachie W. D. The cell cycle of Escherichia coli. Annu Rev Microbiol. 1993;47:199–230. doi: 10.1146/annurev.mi.47.100193.001215. [DOI] [PubMed] [Google Scholar]
- Echols H. Multiple DNA-protein interactions governing high-precision DNA transactions. Science. 1986 Sep 5;233(4768):1050–1056. doi: 10.1126/science.2943018. [DOI] [PubMed] [Google Scholar]
- Elespuru R. K., Yarmolinsky M. B. A colorimetric assay of lysogenic induction designed for screening potential carcinogenic and carcinostatic agents. Environ Mutagen. 1979;1(1):65–78. doi: 10.1002/em.2860010113. [DOI] [PubMed] [Google Scholar]
- Fuller R. S., Funnell B. E., Kornberg A. The dnaA protein complex with the E. coli chromosomal replication origin (oriC) and other DNA sites. Cell. 1984 Oct;38(3):889–900. doi: 10.1016/0092-8674(84)90284-8. [DOI] [PubMed] [Google Scholar]
- Gill D. R., Salmond G. P. The Escherichia coli cell division proteins FtsY, FtsE and FtsX are inner membrane-associated. Mol Gen Genet. 1987 Dec;210(3):504–508. doi: 10.1007/BF00327204. [DOI] [PubMed] [Google Scholar]
- Haniford D. B., Chelouche A. R., Kleckner N. A specific class of IS10 transposase mutants are blocked for target site interactions and promote formation of an excised transposon fragment. Cell. 1989 Oct 20;59(2):385–394. doi: 10.1016/0092-8674(89)90299-7. [DOI] [PubMed] [Google Scholar]
- Hiraga S. Chromosome and plasmid partition in Escherichia coli. Annu Rev Biochem. 1992;61:283–306. doi: 10.1146/annurev.bi.61.070192.001435. [DOI] [PubMed] [Google Scholar]
- Hiraga S., Niki H., Ogura T., Ichinose C., Mori H., Ezaki B., Jaffé A. Chromosome partitioning in Escherichia coli: novel mutants producing anucleate cells. J Bacteriol. 1989 Mar;171(3):1496–1505. doi: 10.1128/jb.171.3.1496-1505.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huisman O., D'Ari R. An inducible DNA replication-cell division coupling mechanism in E. coli. Nature. 1981 Apr 30;290(5809):797–799. doi: 10.1038/290797a0. [DOI] [PubMed] [Google Scholar]
- Isberg R. R., Syvanen M. Compartmentalization of the proteins encoded by IS50R. J Biol Chem. 1985 Mar 25;260(6):3645–3651. [PubMed] [Google Scholar]
- Johnson R. C., Reznikoff W. S. Copy number control of Tn5 transposition. Genetics. 1984 May;107(1):9–18. doi: 10.1093/genetics/107.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson R. C., Reznikoff W. S. Role of the IS50 R proteins in the promotion and control of Tn5 transposition. J Mol Biol. 1984 Aug 25;177(4):645–661. doi: 10.1016/0022-2836(84)90042-1. [DOI] [PubMed] [Google Scholar]
- Kohara Y., Akiyama K., Isono K. The physical map of the whole E. coli chromosome: application of a new strategy for rapid analysis and sorting of a large genomic library. Cell. 1987 Jul 31;50(3):495–508. doi: 10.1016/0092-8674(87)90503-4. [DOI] [PubMed] [Google Scholar]
- Luttinger A. L., Springer A. L., Schmid M. B. A cluster of genes that affects nucleoid segregation in Salmonella typhimurium. New Biol. 1991 Jul;3(7):687–697. [PubMed] [Google Scholar]
- Mizuuchi K. Transpositional recombination: mechanistic insights from studies of mu and other elements. Annu Rev Biochem. 1992;61:1011–1051. doi: 10.1146/annurev.bi.61.070192.005051. [DOI] [PubMed] [Google Scholar]
- Roberts D., Hoopes B. C., McClure W. R., Kleckner N. IS10 transposition is regulated by DNA adenine methylation. Cell. 1985 Nov;43(1):117–130. doi: 10.1016/0092-8674(85)90017-0. [DOI] [PubMed] [Google Scholar]
- Roberts D., Kleckner N. Tn10 transposition promotes RecA-dependent induction of a lambda prophage. Proc Natl Acad Sci U S A. 1988 Aug;85(16):6037–6041. doi: 10.1073/pnas.85.16.6037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts J. W., Roberts C. W., Craig N. L. Escherichia coli recA gene product inactivates phage lambda repressor. Proc Natl Acad Sci U S A. 1978 Oct;75(10):4714–4718. doi: 10.1073/pnas.75.10.4714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothstein S. J., Reznikoff W. S. The functional differences in the inverted repeats of Tn5 are caused by a single base pair nonhomology. Cell. 1981 Jan;23(1):191–199. doi: 10.1016/0092-8674(81)90284-1. [DOI] [PubMed] [Google Scholar]
- Singer M., Baker T. A., Schnitzler G., Deischel S. M., Goel M., Dove W., Jaacks K. J., Grossman A. D., Erickson J. W., Gross C. A. A collection of strains containing genetically linked alternating antibiotic resistance elements for genetic mapping of Escherichia coli. Microbiol Rev. 1989 Mar;53(1):1–24. doi: 10.1128/mr.53.1.1-24.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slater S., Maurer R. Simple phagemid-based system for generating allele replacements in Escherichia coli. J Bacteriol. 1993 Jul;175(13):4260–4262. doi: 10.1128/jb.175.13.4260-4262.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
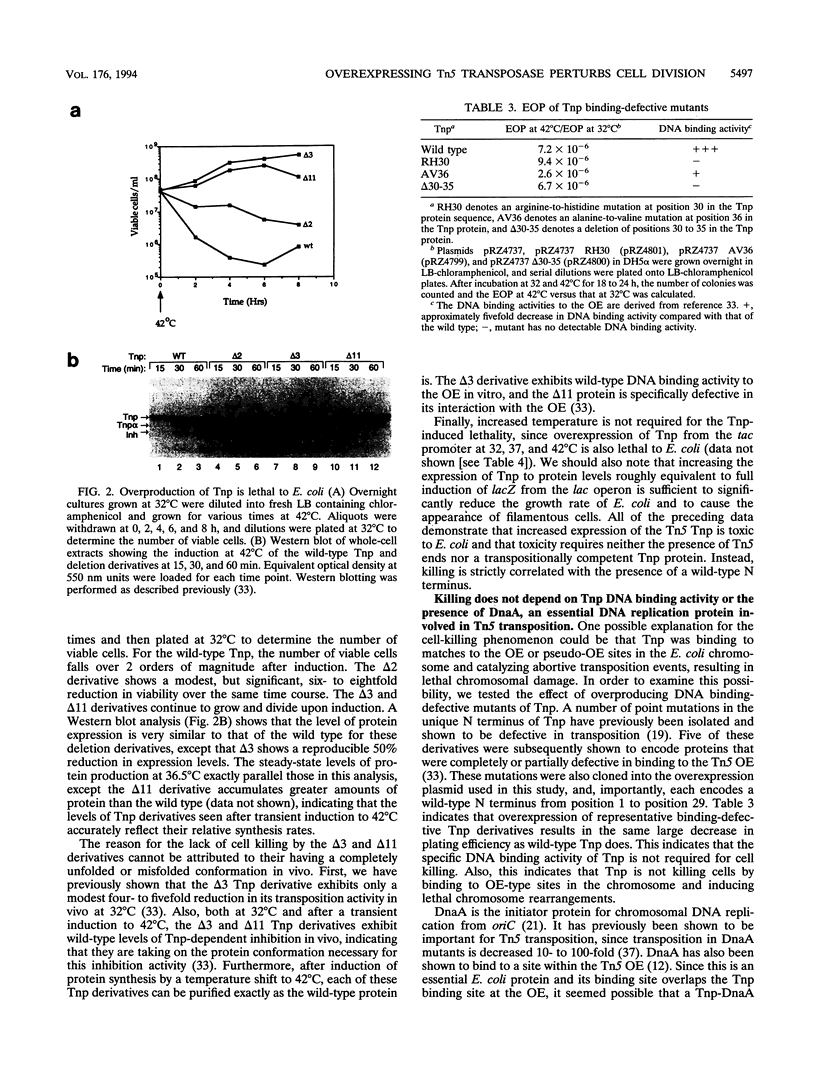
- Wiegand T. W., Reznikoff W. S. Characterization of two hypertransposing Tn5 mutants. J Bacteriol. 1992 Feb;174(4):1229–1239. doi: 10.1128/jb.174.4.1229-1239.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin J. C., Krebs M. P., Reznikoff W. S. Effect of dam methylation on Tn5 transposition. J Mol Biol. 1988 Jan 5;199(1):35–45. doi: 10.1016/0022-2836(88)90377-4. [DOI] [PubMed] [Google Scholar]
- Yin J. C., Reznikoff W. S. dnaA, an essential host gene, and Tn5 transposition. J Bacteriol. 1987 Oct;169(10):4637–4645. doi: 10.1128/jb.169.10.4637-4645.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin J. C., Reznikoff W. S. p2 and inhibition of Tn5 transposition. J Bacteriol. 1988 Jul;170(7):3008–3015. doi: 10.1128/jb.170.7.3008-3015.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Cruz N. B., Weinreich M. D., Wiegand T. W., Krebs M. P., Reznikoff W. S. Characterization of the Tn5 transposase and inhibitor proteins: a model for the inhibition of transposition. J Bacteriol. 1993 Nov;175(21):6932–6938. doi: 10.1128/jb.175.21.6932-6938.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]