Abstract
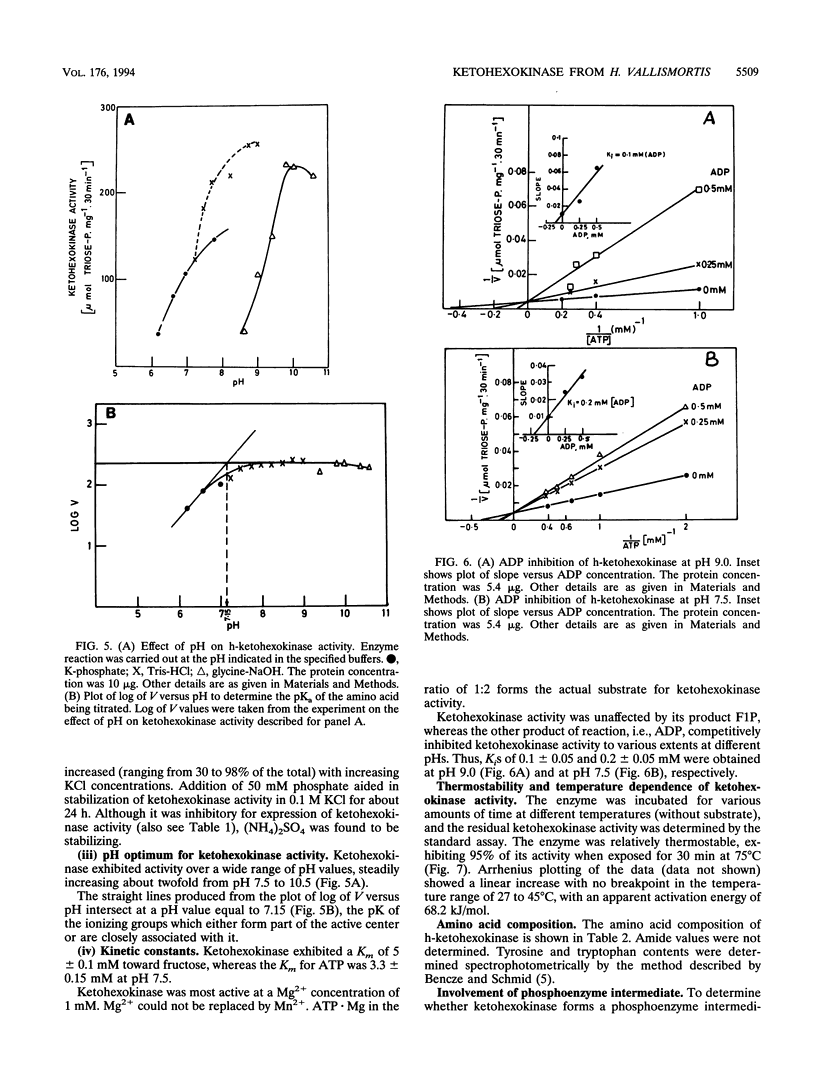
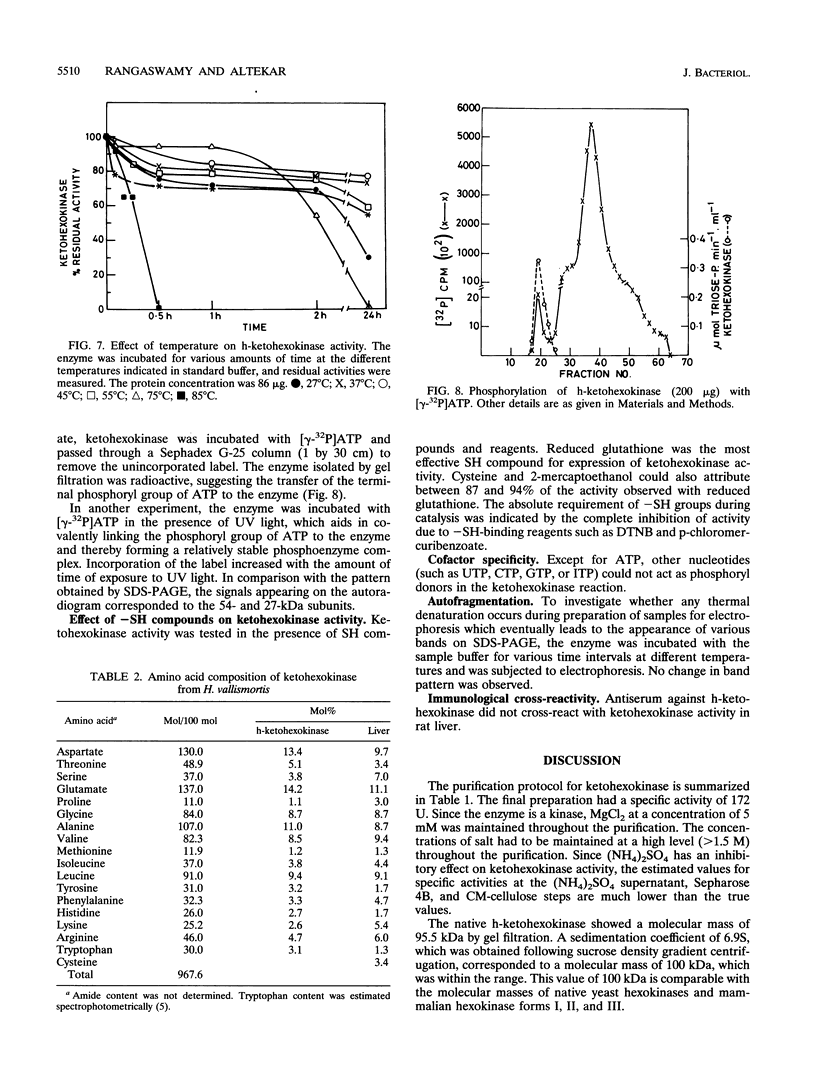
Ketohexokinase (ATP:D-fructose 1-phosphotransferase [EC 2.7.1.3]), detected for the first time in a prokaryote, i.e., the extreme halophile Haloarcula vallismortis, was isolated and characterized from the same archaebacterium. This enzyme was characterized with respect to its molecular mass, amino acid composition, salt dependency, immunological cross-reactivity, and kinetic properties. Gel filtration and sucrose density gradient centrifugation revealed a native molecular mass of 100 kDa for halobacterial ketohexokinase, which is larger than its mammalian counterpart. The enzyme could be labeled by UV irradiation in the presence of [ gamma-32P]ATP, suggesting the involvement of a phosphoenzyme intermediate. Other catalytic features of the enzyme were similar to those of its mammalian counterparts. No antigenic cross-reactivity could be detected between the H. vallismortis ketohexokinase and the ketohexokinases from different rat tissues.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bais R., James H. M., Rofe A. M., Conyers R. A. The purification and properties of human liver ketohexokinase. A role for ketohexokinase and fructose-bisphosphate aldolase in the metabolic production of oxalate from xylitol. Biochem J. 1985 Aug 15;230(1):53–60. doi: 10.1042/bj2300053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonet M. L., Schobert B. The catalytic site is located on subunit I of the ATPase from Halobacterium saccharovorum. A direct photoaffinity labeling study. Eur J Biochem. 1992 Jul 1;207(1):369–376. doi: 10.1111/j.1432-1033.1992.tb17059.x. [DOI] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Chou A. C., Wilson J. E. Purification and properties of rat brain hexokinase. Arch Biochem Biophys. 1972 Jul;151(1):48–55. doi: 10.1016/0003-9861(72)90471-7. [DOI] [PubMed] [Google Scholar]
- Donaldson I. A., Doyle T. C., Matas N. Expression of rat liver ketohexokinase in yeast results in fructose intolerance. Biochem J. 1993 Apr 1;291(Pt 1):179–186. doi: 10.1042/bj2910179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Easterby J. S. The polypeptide chain molecular weight of a mammalian hexokinase. FEBS Lett. 1971 Oct 15;18(1):23–26. doi: 10.1016/0014-5793(71)80397-6. [DOI] [PubMed] [Google Scholar]
- Eisenberg H., Mevarech M., Zaccai G. Biochemical, structural, and molecular genetic aspects of halophilism. Adv Protein Chem. 1992;43:1–62. doi: 10.1016/s0065-3233(08)60553-7. [DOI] [PubMed] [Google Scholar]
- Eley M. H., Burns P. C., Kannapell C. C., Campbell P. S. Cetyltrimethylammonium bromide polyacrylamide gel electrophoresis: estimation of protein subunit molecular weights using cationic detergents. Anal Biochem. 1979 Jan 15;92(2):411–419. doi: 10.1016/0003-2697(79)90679-1. [DOI] [PubMed] [Google Scholar]
- Krishnan G., Altekar W. An unusual class I (Schiff base) fructose-1,6-bisphosphate aldolase from the halophilic archaebacterium Haloarcula vallismortis. Eur J Biochem. 1991 Jan 30;195(2):343–350. doi: 10.1111/j.1432-1033.1991.tb15712.x. [DOI] [PubMed] [Google Scholar]
- Krishnan G., Altekar W. Halophilic class I aldolase and glyceraldehyde-3-phosphate dehydrogenase: some salt-dependent structural features. Biochemistry. 1993 Jan 26;32(3):791–798. doi: 10.1021/bi00054a008. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- MARTIN R. G., AMES B. N. A method for determining the sedimentation behavior of enzymes: application to protein mixtures. J Biol Chem. 1961 May;236:1372–1379. [PubMed] [Google Scholar]
- NAJJAR V. A., McCOY E. E. Mechanism of action of muscle and yeast phosphoglucomutase and a suggested mechanism for yeast hexokinase. Fed Proc. 1958 Dec;17(4):1141–1144. [PubMed] [Google Scholar]
- PARKS R. E., Jr, BEN-GERSHOM E., LARDY H. A. Liver fructokinase. J Biol Chem. 1957 Jul;227(1):231–242. [PubMed] [Google Scholar]
- Paranjpe S. V., Jagannathan V. Properties & kinetics of purified particulate ox heart hexokinase. Indian J Biochem. 1971 Dec;8(4):227–231. [PubMed] [Google Scholar]
- Pringle J. R. The molecular weight of the undegraded polypeptide chain of yeast hexokinase. Biochem Biophys Res Commun. 1970 Apr 8;39(1):46–52. doi: 10.1016/0006-291x(70)90755-2. [DOI] [PubMed] [Google Scholar]
- Raushel F. M., Cleland W. W. Bovine liver fructokinase: purification and kinetic properties. Biochemistry. 1977 May 17;16(10):2169–2175. doi: 10.1021/bi00629a020. [DOI] [PubMed] [Google Scholar]
- Redkar V. D., Kenkare U. W. Bovine brain mitochondrial hexokinase. Solubilization, purification, and role of sulfhydryl residues. J Biol Chem. 1972 Dec 10;247(23):7576–7584. [PubMed] [Google Scholar]
- Saier M. H., Jr, Grenier F. C., Lee C. A., Waygood E. B. Evidence for the evolutionary relatedness of the proteins of the bacterial phosphoenolpyruvate:sugar phosphotransferase system. J Cell Biochem. 1985;27(1):43–56. doi: 10.1002/jcb.240270106. [DOI] [PubMed] [Google Scholar]
- Sánchez J. J., González N. S., Pontis H. G. Fructokinase from rat liver. I. Purification and properties. Biochim Biophys Acta. 1971 Jan 13;227(1):67–78. doi: 10.1016/0005-2744(71)90168-9. [DOI] [PubMed] [Google Scholar]
- Walsh C. T., Jr, Spector L. B. A phosphoenzyme intermediary in phosphoglycerate kinase action. J Biol Chem. 1971 Mar 10;246(5):1255–1261. [PubMed] [Google Scholar]