Abstract
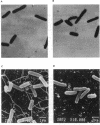
The function of the flagellum-chemotaxis regulon requires the expression of many genes and is positively regulated by the cyclic AMP-catabolite activator protein (cAMP-CAP) complex. In this paper, we show that motile behavior was affected in Escherichia coli hns mutants. The loss of motility resulted from a complete lack of flagella. A decrease in the level of transcription of the flhD and fliA genes, which are both required for the synthesis of flagella, was observed in the presence of an hns mutation. Furthermore, the Fla- phenotype was not reversed to the wild type in the presence of a cfs mutation which renders the flagellum synthesis independent of the cAMP-CAP complex. These results suggest that the H-NS protein acts as a positive regulator of genes involved in the biogenesis of flagella by a mechanism independent of the cAMP-CAP pathway.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adler J. Chemotaxis in bacteria. Science. 1966 Aug 12;153(3737):708–716. doi: 10.1126/science.153.3737.708. [DOI] [PubMed] [Google Scholar]
- Adler J., Templeton B. The effect of environmental conditions on the motility of Escherichia coli. J Gen Microbiol. 1967 Feb;46(2):175–184. doi: 10.1099/00221287-46-2-175. [DOI] [PubMed] [Google Scholar]
- Bartlett D. H., Frantz B. B., Matsumura P. Flagellar transcriptional activators FlbB and FlaI: gene sequences and 5' consensus sequences of operons under FlbB and FlaI control. J Bacteriol. 1988 Apr;170(4):1575–1581. doi: 10.1128/jb.170.4.1575-1581.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertin P., Lejeune P., Colson C., Danchin A. Mutations in bglY, the structural gene for the DNA-binding protein H1 of Escherichia coli, increase the expression of the kanamycin resistance gene carried by plasmid pGR71. Mol Gen Genet. 1992 May;233(1-2):184–192. doi: 10.1007/BF00587578. [DOI] [PubMed] [Google Scholar]
- Bertin P., Lejeune P., Laurent-Winter C., Danchin A. Mutations in bglY, the structural gene for the DNA-binding protein H1, affect expression of several Escherichia coli genes. Biochimie. 1990 Dec;72(12):889–891. doi: 10.1016/0300-9084(90)90008-5. [DOI] [PubMed] [Google Scholar]
- Dersch P., Schmidt K., Bremer E. Synthesis of the Escherichia coli K-12 nucleoid-associated DNA-binding protein H-NS is subjected to growth-phase control and autoregulation. Mol Microbiol. 1993 May;8(5):875–889. doi: 10.1111/j.1365-2958.1993.tb01634.x. [DOI] [PubMed] [Google Scholar]
- Falconi M., Higgins N. P., Spurio R., Pon C. L., Gualerzi C. O. Expression of the gene encoding the major bacterial nucleotide protein H-NS is subject to transcriptional auto-repression. Mol Microbiol. 1993 Oct;10(2):273–282. [PubMed] [Google Scholar]
- Göransson M., Sondén B., Nilsson P., Dagberg B., Forsman K., Emanuelsson K., Uhlin B. E. Transcriptional silencing and thermoregulation of gene expression in Escherichia coli. Nature. 1990 Apr 12;344(6267):682–685. doi: 10.1038/344682a0. [DOI] [PubMed] [Google Scholar]
- Higgins C. F., Hinton J. C., Hulton C. S., Owen-Hughes T., Pavitt G. D., Seirafi A. Protein H1: a role for chromatin structure in the regulation of bacterial gene expression and virulence? Mol Microbiol. 1990 Dec;4(12):2007–2012. doi: 10.1111/j.1365-2958.1990.tb00559.x. [DOI] [PubMed] [Google Scholar]
- Hinton J. C., Santos D. S., Seirafi A., Hulton C. S., Pavitt G. D., Higgins C. F. Expression and mutational analysis of the nucleoid-associated protein H-NS of Salmonella typhimurium. Mol Microbiol. 1992 Aug;6(16):2327–2337. doi: 10.1111/j.1365-2958.1992.tb01408.x. [DOI] [PubMed] [Google Scholar]
- Hitchcock P. J., Brown T. M. Morphological heterogeneity among Salmonella lipopolysaccharide chemotypes in silver-stained polyacrylamide gels. J Bacteriol. 1983 Apr;154(1):269–277. doi: 10.1128/jb.154.1.269-277.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulton C. S., Seirafi A., Hinton J. C., Sidebotham J. M., Waddell L., Pavitt G. D., Owen-Hughes T., Spassky A., Buc H., Higgins C. F. Histone-like protein H1 (H-NS), DNA supercoiling, and gene expression in bacteria. Cell. 1990 Nov 2;63(3):631–642. doi: 10.1016/0092-8674(90)90458-q. [DOI] [PubMed] [Google Scholar]
- Jacquet M., Cukier-Kahn R., Pla J., Gros F. A thermostable protein factor acting on in vitro DNA transcription. Biochem Biophys Res Commun. 1971 Dec 17;45(6):1597–1607. doi: 10.1016/0006-291x(71)90204-x. [DOI] [PubMed] [Google Scholar]
- Jones C. J., Aizawa S. The bacterial flagellum and flagellar motor: structure, assembly and function. Adv Microb Physiol. 1991;32:109–172. doi: 10.1016/s0065-2911(08)60007-7. [DOI] [PubMed] [Google Scholar]
- Komeda Y., Icho T., Iino T. Effects of galU mutation on flagellar formation in Escherichia coli. J Bacteriol. 1977 Feb;129(2):908–915. doi: 10.1128/jb.129.2.908-915.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komeda Y. Transcriptional control of flagellar genes in Escherichia coli K-12. J Bacteriol. 1986 Dec;168(3):1315–1318. doi: 10.1128/jb.168.3.1315-1318.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Teana A., Brandi A., Falconi M., Spurio R., Pon C. L., Gualerzi C. O. Identification of a cold shock transcriptional enhancer of the Escherichia coli gene encoding nucleoid protein H-NS. Proc Natl Acad Sci U S A. 1991 Dec 1;88(23):10907–10911. doi: 10.1073/pnas.88.23.10907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lejeune P., Bertin P., Walon C., Willemot K., Colson C., Danchin A. A locus involved in kanamycin, chloramphenicol and L-serine resistance is located in the bglY-galU region of the Escherichia coli K12 chromosome. Mol Gen Genet. 1989 Aug;218(2):361–363. doi: 10.1007/BF00331292. [DOI] [PubMed] [Google Scholar]
- Lejeune P., Danchin A. Mutations in the bglY gene increase the frequency of spontaneous deletions in Escherichia coli K-12. Proc Natl Acad Sci U S A. 1990 Jan;87(1):360–363. doi: 10.1073/pnas.87.1.360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macnab R. M. Genetics and biogenesis of bacterial flagella. Annu Rev Genet. 1992;26:131–158. doi: 10.1146/annurev.ge.26.120192.001023. [DOI] [PubMed] [Google Scholar]
- May G., Dersch P., Haardt M., Middendorf A., Bremer E. The osmZ (bglY) gene encodes the DNA-binding protein H-NS (H1a), a component of the Escherichia coli K12 nucleoid. Mol Gen Genet. 1990 Oct;224(1):81–90. doi: 10.1007/BF00259454. [DOI] [PubMed] [Google Scholar]
- Parker C. T., Kloser A. W., Schnaitman C. A., Stein M. A., Gottesman S., Gibson B. W. Role of the rfaG and rfaP genes in determining the lipopolysaccharide core structure and cell surface properties of Escherichia coli K-12. J Bacteriol. 1992 Apr;174(8):2525–2538. doi: 10.1128/jb.174.8.2525-2538.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi W., Bogdanov M., Dowhan W., Zusman D. R. The pss and psd genes are required for motility and chemotaxis in Escherichia coli. J Bacteriol. 1993 Dec;175(23):7711–7714. doi: 10.1128/jb.175.23.7711-7714.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi W., Li C., Louise C. J., Adler J. Mechanism of adverse conditions causing lack of flagella in Escherichia coli. J Bacteriol. 1993 Apr;175(8):2236–2240. doi: 10.1128/jb.175.8.2236-2240.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi W., Zhou Y., Wild J., Adler J., Gross C. A. DnaK, DnaJ, and GrpE are required for flagellum synthesis in Escherichia coli. J Bacteriol. 1992 Oct;174(19):6256–6263. doi: 10.1128/jb.174.19.6256-6263.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueguchi C., Kakeda M., Mizuno T. Autoregulatory expression of the Escherichia coli hns gene encoding a nucleoid protein: H-NS functions as a repressor of its own transcription. Mol Gen Genet. 1993 Jan;236(2-3):171–178. doi: 10.1007/BF00277109. [DOI] [PubMed] [Google Scholar]
- VOGEL H. J., BONNER D. M. Acetylornithinase of Escherichia coli: partial purification and some properties. J Biol Chem. 1956 Jan;218(1):97–106. [PubMed] [Google Scholar]
- Vogler A. P., Lengeler J. W. Indirect role of adenylate cyclase and cyclic AMP in chemotaxis to phosphotransferase system carbohydrates in Escherichia coli K-12. J Bacteriol. 1987 Feb;169(2):593–599. doi: 10.1128/jb.169.2.593-599.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada H., Yoshida T., Tanaka K., Sasakawa C., Mizuno T. Molecular analysis of the Escherichia coli hns gene encoding a DNA-binding protein, which preferentially recognizes curved DNA sequences. Mol Gen Genet. 1991 Nov;230(1-2):332–336. doi: 10.1007/BF00290685. [DOI] [PubMed] [Google Scholar]