Abstract
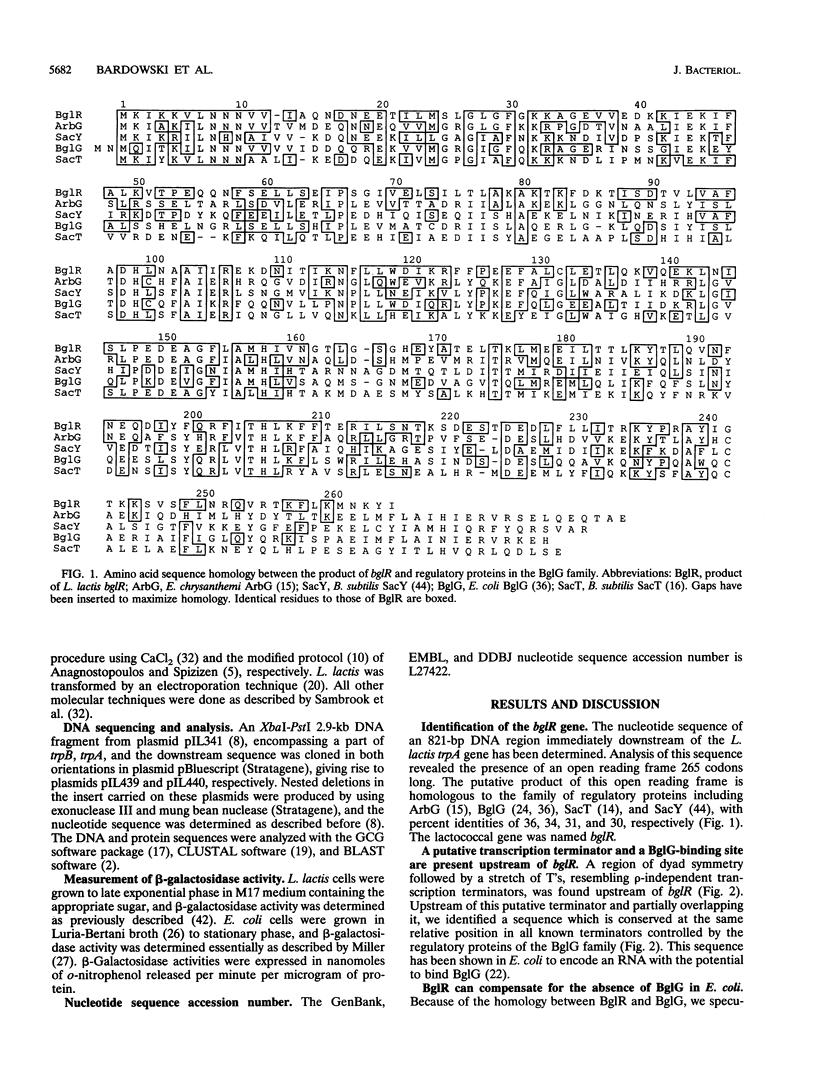
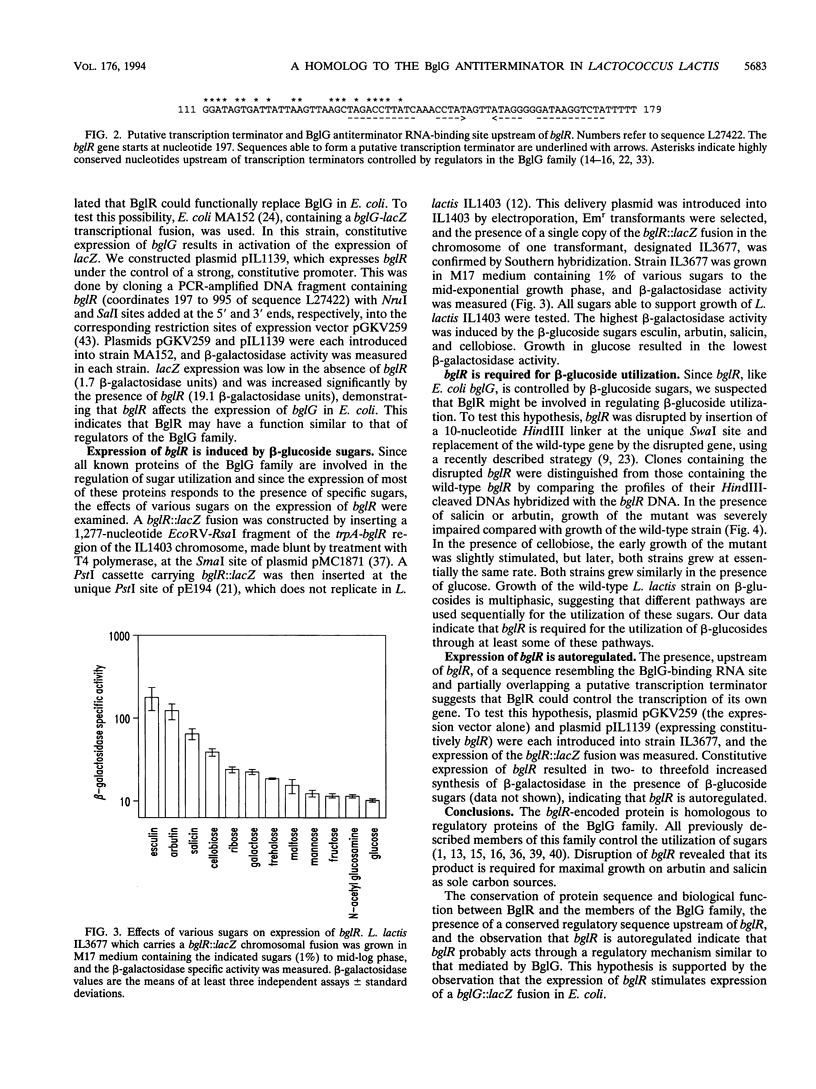
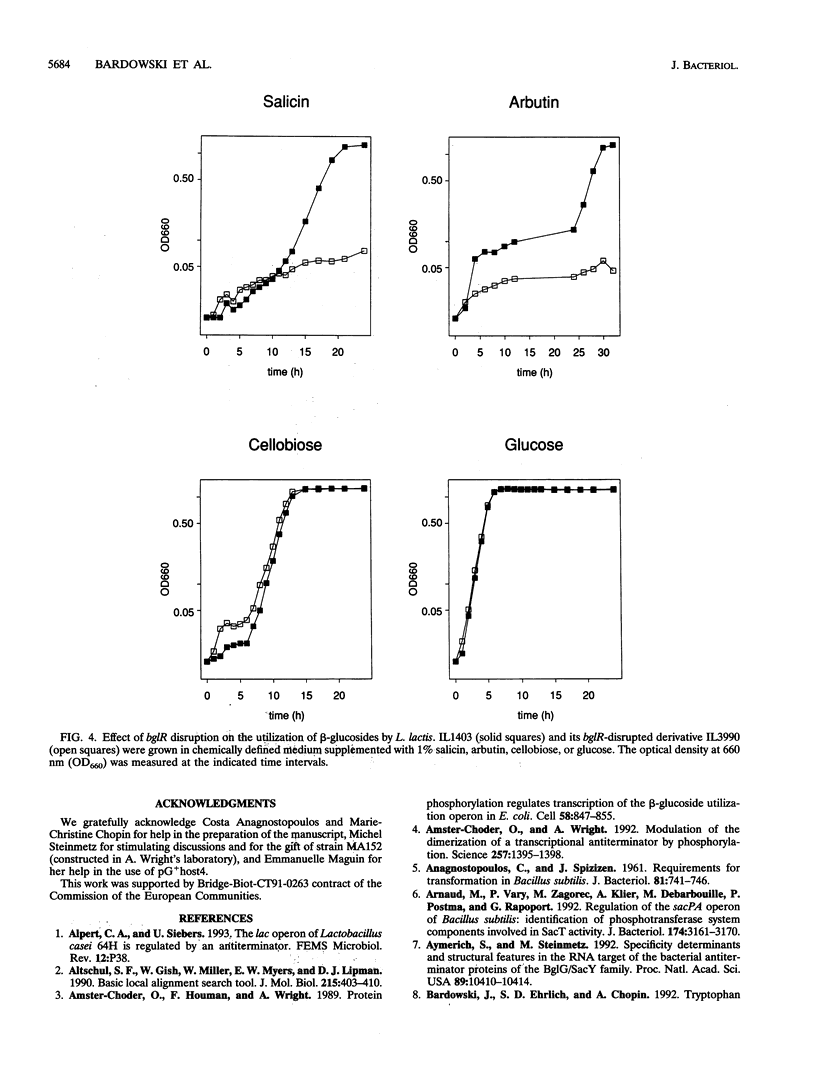
A fragment of the Lactococcus lactis chromosome containing an open reading frame of 265 codons, denoted bglR, has been characterized. The polypeptide encoded by bglR shares 36 to 30% sequence identity with a family of regulatory proteins including ArbG from Erwinia chrysanthemi, BglG from Escherichia coli, and SacT and SacY from Bacillus subtilis. These regulatory proteins are involved in positive control of the utilization of different sugars by transcription antitermination. For some of these regulatory proteins it has been demonstrated that antitermination is exerted by binding to a conserved RNA sequence, partially overlapping the transcription terminator and thus preventing transcription termination. Upstream of bglR, we identified a transcription terminator whose 5' end was overlapped by a 32-bp sequence, highly homologous to the RNA-binding site that is conserved in other regulatory systems. Constitutive expression of bglR in E. coli increased the expression of a bglG::lacZ transcriptional fusion. The fact that that the expression of BglG is autoregulated in E. coli suggests that BglG and BglR are functionally equivalent. In L. lactis, we observed that (i) the expression of a bglR::lacZ fusion is increased by beta-glucoside sugars, (ii) disruption of bglR impairs growth on some beta-glucosides, and (iii) the expression of bglR is positively autoregulated. Because of these structural and functional similarities between BglR and the transcription antiterminators of the BglG family, we propose that BglR may be the lactococcal counterpart of the E. coli BglG regulator of beta-glucoside utilization.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. Basic local alignment search tool. J Mol Biol. 1990 Oct 5;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Amster-Choder O., Houman F., Wright A. Protein phosphorylation regulates transcription of the beta-glucoside utilization operon in E. coli. Cell. 1989 Sep 8;58(5):847–855. doi: 10.1016/0092-8674(89)90937-9. [DOI] [PubMed] [Google Scholar]
- Amster-Choder O., Wright A. Modulation of the dimerization of a transcriptional antiterminator protein by phosphorylation. Science. 1992 Sep 4;257(5075):1395–1398. doi: 10.1126/science.1382312. [DOI] [PubMed] [Google Scholar]
- Anagnostopoulos C., Spizizen J. REQUIREMENTS FOR TRANSFORMATION IN BACILLUS SUBTILIS. J Bacteriol. 1961 May;81(5):741–746. doi: 10.1128/jb.81.5.741-746.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnaud M., Vary P., Zagorec M., Klier A., Debarbouille M., Postma P., Rapoport G. Regulation of the sacPA operon of Bacillus subtilis: identification of phosphotransferase system components involved in SacT activity. J Bacteriol. 1992 May;174(10):3161–3170. doi: 10.1128/jb.174.10.3161-3170.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aymerich S., Steinmetz M. Specificity determinants and structural features in the RNA target of the bacterial antiterminator proteins of the BglG/SacY family. Proc Natl Acad Sci U S A. 1992 Nov 1;89(21):10410–10414. doi: 10.1073/pnas.89.21.10410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardowski J., Ehrlich S. D., Chopin A. Tryptophan biosynthesis genes in Lactococcus lactis subsp. lactis. J Bacteriol. 1992 Oct;174(20):6563–6570. doi: 10.1128/jb.174.20.6563-6570.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas I., Gruss A., Ehrlich S. D., Maguin E. High-efficiency gene inactivation and replacement system for gram-positive bacteria. J Bacteriol. 1993 Jun;175(11):3628–3635. doi: 10.1128/jb.175.11.3628-3635.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chopin A., Chopin M. C., Moillo-Batt A., Langella P. Two plasmid-determined restriction and modification systems in Streptococcus lactis. Plasmid. 1984 May;11(3):260–263. doi: 10.1016/0147-619x(84)90033-7. [DOI] [PubMed] [Google Scholar]
- Chopin M. C., Chopin A., Rouault A., Galleron N. Insertion and amplification of foreign genes in the Lactococcus lactis subsp. lactis chromosome. Appl Environ Microbiol. 1989 Jul;55(7):1769–1774. doi: 10.1128/aem.55.7.1769-1774.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crutz A. M., Steinmetz M., Aymerich S., Richter R., Le Coq D. Induction of levansucrase in Bacillus subtilis: an antitermination mechanism negatively controlled by the phosphotransferase system. J Bacteriol. 1990 Feb;172(2):1043–1050. doi: 10.1128/jb.172.2.1043-1050.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debarbouille M., Arnaud M., Fouet A., Klier A., Rapoport G. The sacT gene regulating the sacPA operon in Bacillus subtilis shares strong homology with transcriptional antiterminators. J Bacteriol. 1990 Jul;172(7):3966–3973. doi: 10.1128/jb.172.7.3966-3973.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouet A., Arnaud M., Klier A., Rapoport G. Bacillus subtilis sucrose-specific enzyme II of the phosphotransferase system: expression in Escherichia coli and homology to enzymes II from enteric bacteria. Proc Natl Acad Sci U S A. 1987 Dec;84(24):8773–8777. doi: 10.1073/pnas.84.24.8773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins D. G., Sharp P. M. CLUSTAL: a package for performing multiple sequence alignment on a microcomputer. Gene. 1988 Dec 15;73(1):237–244. doi: 10.1016/0378-1119(88)90330-7. [DOI] [PubMed] [Google Scholar]
- Holo H., Nes I. F. High-Frequency Transformation, by Electroporation, of Lactococcus lactis subsp. cremoris Grown with Glycine in Osmotically Stabilized Media. Appl Environ Microbiol. 1989 Dec;55(12):3119–3123. doi: 10.1128/aem.55.12.3119-3123.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horinouchi S., Weisblum B. Nucleotide sequence and functional map of pE194, a plasmid that specifies inducible resistance to macrolide, lincosamide, and streptogramin type B antibodies. J Bacteriol. 1982 May;150(2):804–814. doi: 10.1128/jb.150.2.804-814.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houman F., Diaz-Torres M. R., Wright A. Transcriptional antitermination in the bgl operon of E. coli is modulated by a specific RNA binding protein. Cell. 1990 Sep 21;62(6):1153–1163. doi: 10.1016/0092-8674(90)90392-r. [DOI] [PubMed] [Google Scholar]
- Maguin E., Duwat P., Hege T., Ehrlich D., Gruss A. New thermosensitive plasmid for gram-positive bacteria. J Bacteriol. 1992 Sep;174(17):5633–5638. doi: 10.1128/jb.174.17.5633-5638.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahadevan S., Reynolds A. E., Wright A. Positive and negative regulation of the bgl operon in Escherichia coli. J Bacteriol. 1987 Jun;169(6):2570–2578. doi: 10.1128/jb.169.6.2570-2578.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahadevan S., Wright A. A bacterial gene involved in transcription antitermination: regulation at a rho-independent terminator in the bgl operon of E. coli. Cell. 1987 Jul 31;50(3):485–494. doi: 10.1016/0092-8674(87)90502-2. [DOI] [PubMed] [Google Scholar]
- Poolman B., Konings W. N. Relation of growth of Streptococcus lactis and Streptococcus cremoris to amino acid transport. J Bacteriol. 1988 Feb;170(2):700–707. doi: 10.1128/jb.170.2.700-707.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds A. E., Felton J., Wright A. Insertion of DNA activates the cryptic bgl operon in E. coli K12. Nature. 1981 Oct 22;293(5834):625–629. doi: 10.1038/293625a0. [DOI] [PubMed] [Google Scholar]
- Reynolds A. E., Mahadevan S., LeGrice S. F., Wright A. Enhancement of bacterial gene expression by insertion elements or by mutation in a CAP-cAMP binding site. J Mol Biol. 1986 Sep 5;191(1):85–95. doi: 10.1016/0022-2836(86)90424-9. [DOI] [PubMed] [Google Scholar]
- Schnetz K., Rak B. Beta-glucoside permease represses the bgl operon of Escherichia coli by phosphorylation of the antiterminator protein and also interacts with glucose-specific enzyme III, the key element in catabolite control. Proc Natl Acad Sci U S A. 1990 Jul;87(13):5074–5078. doi: 10.1073/pnas.87.13.5074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnetz K., Rak B. IS5: a mobile enhancer of transcription in Escherichia coli. Proc Natl Acad Sci U S A. 1992 Feb 15;89(4):1244–1248. doi: 10.1073/pnas.89.4.1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnetz K., Rak B. Regulation of the bgl operon of Escherichia coli by transcriptional antitermination. EMBO J. 1988 Oct;7(10):3271–3277. doi: 10.1002/j.1460-2075.1988.tb03194.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnetz K., Toloczyki C., Rak B. Beta-glucoside (bgl) operon of Escherichia coli K-12: nucleotide sequence, genetic organization, and possible evolutionary relationship to regulatory components of two Bacillus subtilis genes. J Bacteriol. 1987 Jun;169(6):2579–2590. doi: 10.1128/jb.169.6.2579-2590.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapira S. K., Chou J., Richaud F. V., Casadaban M. J. New versatile plasmid vectors for expression of hybrid proteins coded by a cloned gene fused to lacZ gene sequences encoding an enzymatically active carboxy-terminal portion of beta-galactosidase. Gene. 1983 Nov;25(1):71–82. doi: 10.1016/0378-1119(83)90169-5. [DOI] [PubMed] [Google Scholar]
- Smid E. J., Konings W. N. Relationship between utilization of proline and proline-containing peptides and growth of Lactococcus lactis. J Bacteriol. 1990 Sep;172(9):5286–5292. doi: 10.1128/jb.172.9.5286-5292.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmetz M., Le Coq D., Aymerich S. Induction of saccharolytic enzymes by sucrose in Bacillus subtilis: evidence for two partially interchangeable regulatory pathways. J Bacteriol. 1989 Mar;171(3):1519–1523. doi: 10.1128/jb.171.3.1519-1523.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T. Restriction of plasmid-mediated transformation in Bacillus subtilis 168. Mol Gen Genet. 1979 Sep;175(2):235–237. doi: 10.1007/BF00425542. [DOI] [PubMed] [Google Scholar]
- Zukowski M. M., Miller L., Cosgwell P., Chen K., Aymerich S., Steinmetz M. Nucleotide sequence of the sacS locus of Bacillus subtilis reveals the presence of two regulatory genes. Gene. 1990 May 31;90(1):153–155. doi: 10.1016/0378-1119(90)90453-x. [DOI] [PubMed] [Google Scholar]
- el Hassouni M., Henrissat B., Chippaux M., Barras F. Nucleotide sequences of the arb genes, which control beta-glucoside utilization in Erwinia chrysanthemi: comparison with the Escherichia coli bgl operon and evidence for a new beta-glycohydrolase family including enzymes from eubacteria, archeabacteria, and humans. J Bacteriol. 1992 Feb;174(3):765–777. doi: 10.1128/jb.174.3.765-777.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Guchte M., Kok J., Venema G. Distance-dependent translational coupling and interference in Lactococcus lactis. Mol Gen Genet. 1991 May;227(1):65–71. doi: 10.1007/BF00260708. [DOI] [PubMed] [Google Scholar]
- van der Vossen J. M., van der Lelie D., Venema G. Isolation and characterization of Streptococcus cremoris Wg2-specific promoters. Appl Environ Microbiol. 1987 Oct;53(10):2452–2457. doi: 10.1128/aem.53.10.2452-2457.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]



