Abstract
An in vitro system based on extracts of Escherichia coli infected with bacteriophage T7 was used to study genetic deletions between directly repeated sequences. The frequency of deletion was highest under conditions in which the DNA was actively replicating. Deletion frequency increased markedly with the length of the direct repeat both in vitro and in vivo. When a T7 gene was interrupted by 93 bp of nonsense sequence flanked by 20-bp direct repeats, the region between the repeats was deleted in about 1 out of every 1,600 genomes during each round of replication. Very similar values were found for deletion frequency in vivo and in vitro. The deletion frequency was essentially unaffected by a recA mutation in the host. When a double-strand break was placed between the repeats, repair of this strand break was often accompanied by the deletion of the DNA between the direct repeats, suggesting that break rejoining could contribute to deletion during in vitro DNA replication.
Full text
PDF
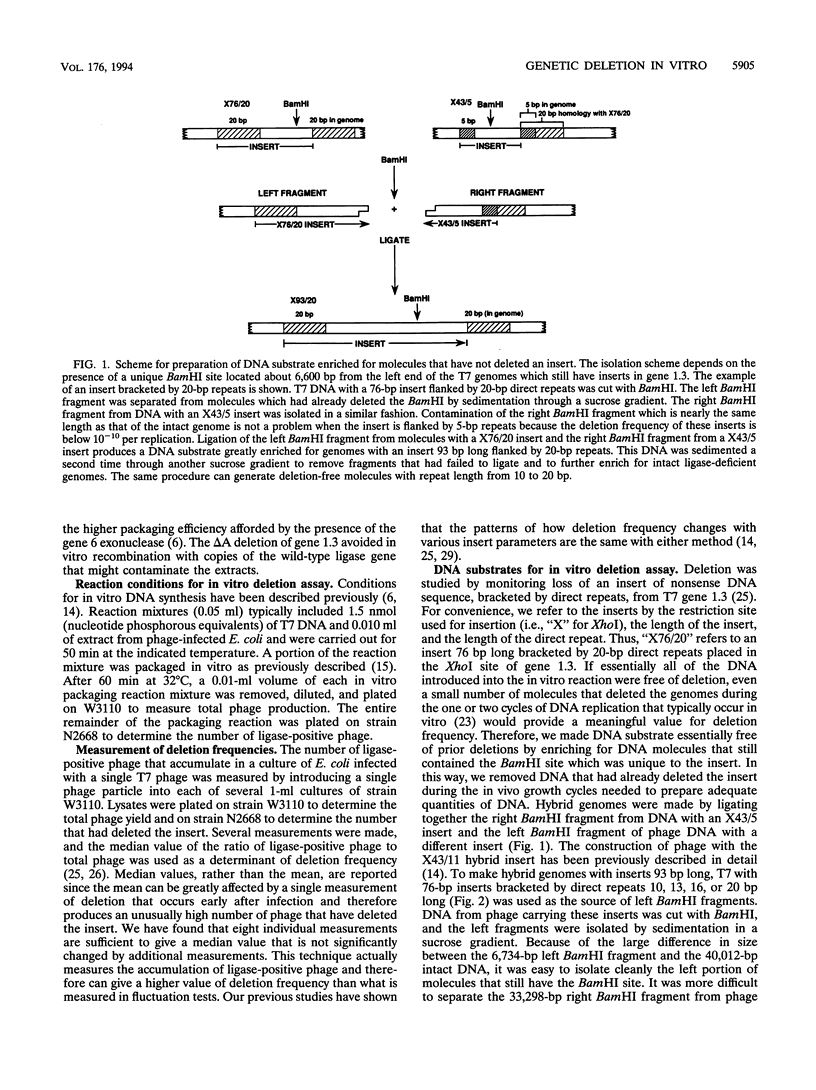


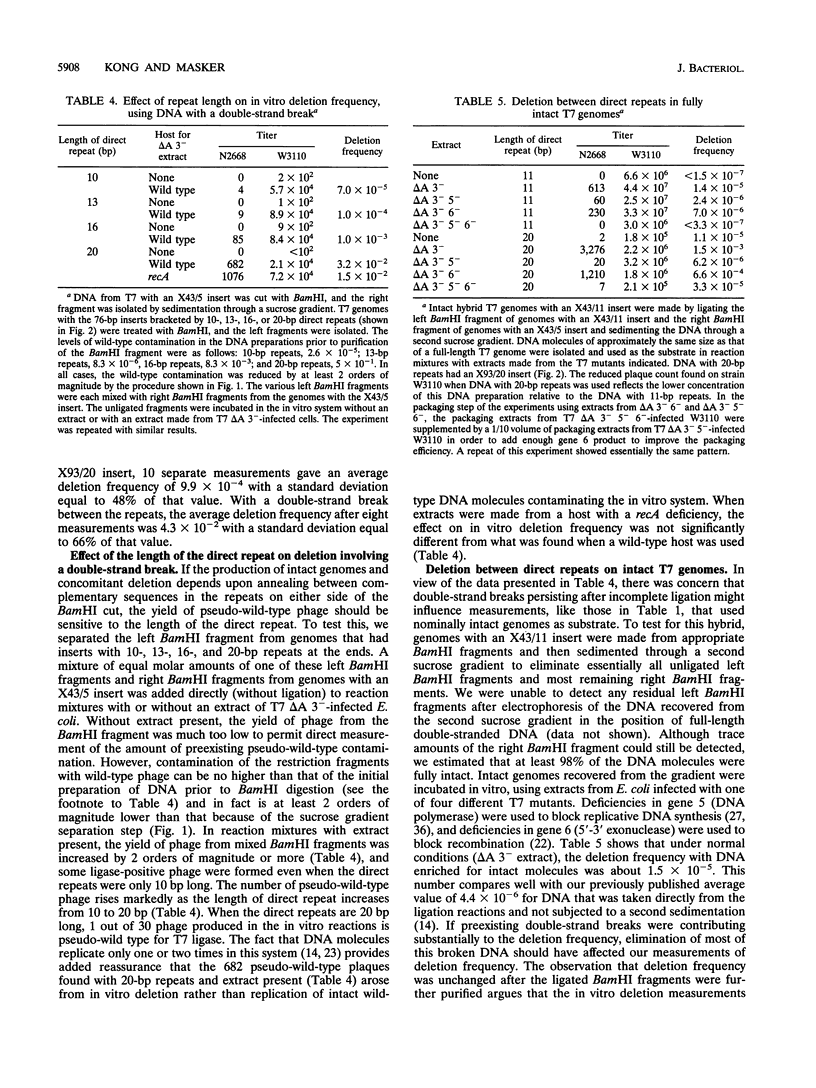



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Albertini A. M., Hofer M., Calos M. P., Miller J. H. On the formation of spontaneous deletions: the importance of short sequence homologies in the generation of large deletions. Cell. 1982 Jun;29(2):319–328. doi: 10.1016/0092-8674(82)90148-9. [DOI] [PubMed] [Google Scholar]
- Balbinder E., Mac Vean C., Williams R. E. Overlapping direct repeats stimulate deletions in specially designed derivatives of plasmid pBR325 in Escherichia coli. Mutat Res. 1989 Oct;214(2):233–252. doi: 10.1016/0027-5107(89)90168-1. [DOI] [PubMed] [Google Scholar]
- Conley E. C., Saunders V. A., Saunders J. R. Deletion and rearrangement of plasmid DNA during transformation of Escherichia coli with linear plasmid molecules. Nucleic Acids Res. 1986 Nov 25;14(22):8905–8917. doi: 10.1093/nar/14.22.8905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DasGupta U., Weston-Hafer K., Berg D. E. Local DNA sequence control of deletion formation in Escherichia coli plasmid pBR322. Genetics. 1987 Jan;115(1):41–49. doi: 10.1093/genetics/115.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodson L. A., Foote R. S., Mitra S., Masker W. E. Mutagenesis of bacteriophage T7 in vitro by incorporation of O6-methylguanine during DNA synthesis. Proc Natl Acad Sci U S A. 1982 Dec;79(23):7440–7444. doi: 10.1073/pnas.79.23.7440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn J. J., Studier F. W. Complete nucleotide sequence of bacteriophage T7 DNA and the locations of T7 genetic elements. J Mol Biol. 1983 Jun 5;166(4):477–535. doi: 10.1016/s0022-2836(83)80282-4. [DOI] [PubMed] [Google Scholar]
- Ehrlich S. D., Bierne H., d'Alençon E., Vilette D., Petranovic M., Noirot P., Michel B. Mechanisms of illegitimate recombination. Gene. 1993 Dec 15;135(1-2):161–166. doi: 10.1016/0378-1119(93)90061-7. [DOI] [PubMed] [Google Scholar]
- Farabaugh P. J., Schmeissner U., Hofer M., Miller J. H. Genetic studies of the lac repressor. VII. On the molecular nature of spontaneous hotspots in the lacI gene of Escherichia coli. J Mol Biol. 1978 Dec 25;126(4):847–857. doi: 10.1016/0022-2836(78)90023-2. [DOI] [PubMed] [Google Scholar]
- Fishman-Lobell J., Haber J. E. Removal of nonhomologous DNA ends in double-strand break recombination: the role of the yeast ultraviolet repair gene RAD1. Science. 1992 Oct 16;258(5081):480–484. doi: 10.1126/science.1411547. [DOI] [PubMed] [Google Scholar]
- Ganesh A., North P., Thacker J. Repair and misrepair of site-specific DNA double-strand breaks by human cell extracts. Mutat Res. 1993 May;299(3-4):251–259. doi: 10.1016/0165-1218(93)90101-i. [DOI] [PubMed] [Google Scholar]
- Ikeda H., Aoki K., Naito A. Illegitimate recombination mediated in vitro by DNA gyrase of Escherichia coli: structure of recombinant DNA molecules. Proc Natl Acad Sci U S A. 1982 Jun;79(12):3724–3728. doi: 10.1073/pnas.79.12.3724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong D., Masker W. Deletion between directly repeated DNA sequences measured in extracts of bacteriophage T7-infected Escherichia coli. J Biol Chem. 1993 Apr 15;268(11):7721–7727. [PubMed] [Google Scholar]
- Kuemmerle N. B., Masker W. E. In vitro packaging of UV radiation-damaged DNA from bacteriophage T7. J Virol. 1977 Sep;23(3):509–516. doi: 10.1128/jvi.23.3.509-516.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel T. A. Misalignment-mediated DNA synthesis errors. Biochemistry. 1990 Sep 4;29(35):8003–8011. doi: 10.1021/bi00487a001. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A., Soni A. Mutagenesis by transient misalignment. J Biol Chem. 1988 Oct 15;263(29):14784–14789. [PubMed] [Google Scholar]
- Lin F. L., Sperle K., Sternberg N. Model for homologous recombination during transfer of DNA into mouse L cells: role for DNA ends in the recombination process. Mol Cell Biol. 1984 Jun;4(6):1020–1034. doi: 10.1128/mcb.4.6.1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovett S. T., Drapkin P. T., Sutera V. A., Jr, Gluckman-Peskind T. J. A sister-strand exchange mechanism for recA-independent deletion of repeated DNA sequences in Escherichia coli. Genetics. 1993 Nov;135(3):631–642. doi: 10.1093/genetics/135.3.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masamune Y., Frenkel G. D., Richardson C. C. A mutant of bacteriophage T7 deficient in polynucleotide ligase. J Biol Chem. 1971 Nov 25;246(22):6874–6879. [PubMed] [Google Scholar]
- Masker W. E., Kuemmerle N. B., Allison D. P. In vitro packaging of bacteriophate T7 DNA synthesized in vitro. J Virol. 1978 Jul;27(1):149–163. doi: 10.1128/jvi.27.1.149-163.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masker W. E., Kuemmerle N. B. In vitro recombination of bacteriophage T7 DNA damaged by UV radiation. J Virol. 1980 Jan;33(1):330–339. doi: 10.1128/jvi.33.1.330-339.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masker W. In vitro repair of double-strand breaks accompanied by recombination in bacteriophage T7 DNA. J Bacteriol. 1992 Jan;174(1):155–160. doi: 10.1128/jb.174.1.155-160.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pachl C., Schubach W., Eisenman R., Linial M. Expression of c-myc RNA in bursal lymphoma cell lines: identification of c-myc-encoded proteins by hybrid-selected translation. Cell. 1983 Jun;33(2):335–344. doi: 10.1016/0092-8674(83)90415-4. [DOI] [PubMed] [Google Scholar]
- Pierce J. C., Kong D., Masker W. The effect of the length of direct repeats and the presence of palindromes on deletion between directly repeated DNA sequences in bacteriophage T7. Nucleic Acids Res. 1991 Jul 25;19(14):3901–3905. doi: 10.1093/nar/19.14.3901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce J. C., Masker W. Genetic deletions between directly repeated sequences in bacteriophage T7. Mol Gen Genet. 1989 Jun;217(2-3):215–222. doi: 10.1007/BF02464884. [DOI] [PubMed] [Google Scholar]
- Scearce L. M., Masker W. Deletion between direct repeats in bacteriophage T7 gene 1.2. Mutat Res. 1993 Aug;288(2):301–310. doi: 10.1016/0027-5107(93)90098-z. [DOI] [PubMed] [Google Scholar]
- Scearce L. M., Pierce J. C., McInroy B., Masker W. Deletion mutagenesis independent of recombination in bacteriophage T7. J Bacteriol. 1991 Jan;173(2):869–878. doi: 10.1128/jb.173.2.869-878.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaaper R. M., Danforth B. N., Glickman B. W. Mechanisms of spontaneous mutagenesis: an analysis of the spectrum of spontaneous mutation in the Escherichia coli lacI gene. J Mol Biol. 1986 May 20;189(2):273–284. doi: 10.1016/0022-2836(86)90509-7. [DOI] [PubMed] [Google Scholar]
- Schiestl R. H., Igarashi S., Hastings P. J. Analysis of the mechanism for reversion of a disrupted gene. Genetics. 1988 Jun;119(2):237–247. doi: 10.1093/genetics/119.2.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulte-Frohlinde D., Worm K. H., Merz M. Double-strand breaks in plasmid DNA and the induction of deletions. Mutat Res. 1993 May;299(3-4):233–250. doi: 10.1016/0165-1218(93)90100-r. [DOI] [PubMed] [Google Scholar]
- Singer B. S., Westlye J. Deletion formation in bacteriophage T4. J Mol Biol. 1988 Jul 20;202(2):233–243. doi: 10.1016/0022-2836(88)90454-8. [DOI] [PubMed] [Google Scholar]
- Streisinger G., Okada Y., Emrich J., Newton J., Tsugita A., Terzaghi E., Inouye M. Frameshift mutations and the genetic code. This paper is dedicated to Professor Theodosius Dobzhansky on the occasion of his 66th birthday. Cold Spring Harb Symp Quant Biol. 1966;31:77–84. doi: 10.1101/sqb.1966.031.01.014. [DOI] [PubMed] [Google Scholar]
- Studier F. W. Bacteriophage T7. Science. 1972 Apr 28;176(4033):367–376. doi: 10.1126/science.176.4033.367. [DOI] [PubMed] [Google Scholar]
- Studier F. W. Genetic analysis of non-essential bacteriophage T7 genes. J Mol Biol. 1973 Sep 15;79(2):227–236. doi: 10.1016/0022-2836(73)90002-8. [DOI] [PubMed] [Google Scholar]
- Studier F. W. The genetics and physiology of bacteriophage T7. Virology. 1969 Nov;39(3):562–574. doi: 10.1016/0042-6822(69)90104-4. [DOI] [PubMed] [Google Scholar]
- Thacker J., Chalk J., Ganesh A., North P. A mechanism for deletion formation in DNA by human cell extracts: the involvement of short sequence repeats. Nucleic Acids Res. 1992 Dec 11;20(23):6183–6188. doi: 10.1093/nar/20.23.6183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinh T. Q., Sinden R. R. Preferential DNA secondary structure mutagenesis in the lagging strand of replication in E. coli. Nature. 1991 Aug 8;352(6335):544–547. doi: 10.1038/352544a0. [DOI] [PubMed] [Google Scholar]
- Weston-Hafer K., Berg D. E. Deletions in plasmid pBR322: replication slippage involving leading and lagging strands. Genetics. 1991 Apr;127(4):649–655. doi: 10.1093/genetics/127.4.649. [DOI] [PMC free article] [PubMed] [Google Scholar]



