Abstract
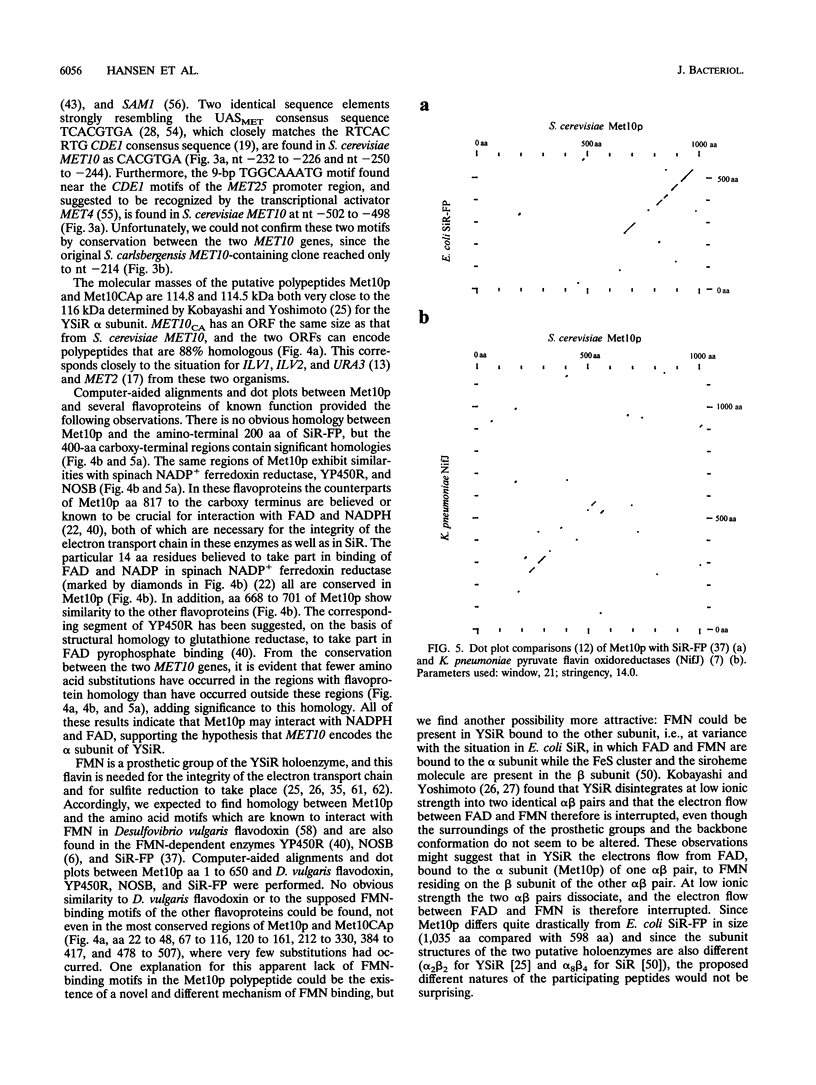
The yeast assimilatory sulfate reductase is a complex enzyme that is responsible for conversion of sulfite into sulfide. To obtain information on the nature of this enzyme, we isolated and sequenced the MET10 gene of Saccharomyces cerevisiae and a divergent MET10 allele from Saccharomyces carlsbergensis. The polypeptides deduced from the identically sized open reading frames (1,035 amino acids) of both MET10 genes have molecular masses of around 115 kDa and are 88% identical to each other. The transcript of S. cerevisiae MET10 has a size comparable to that of the open reading frame and is transcriptionally repressed by methionine in a way similar to that seen for other MET genes of S. cerevisiae. Distinct homology was found between the putative MET10-encoded polypeptide and flavin-interacting parts of the sulfite reductase flavoprotein subunit (encoded by cysJ) from Escherichia coli and several other flavoproteins. A significant N-terminal homology to pyruvate flavodoxin oxidoreductase (encoded by nifJ) from Klebsiella pneumoniae, together with a lack of obvious flavin mononucleotide-binding motifs in the MET10 deduced amino acid sequence, suggests that the yeast assimilatory sulfite reductase is a distinct type of sulfite reductase.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arndt K., Fink G. R. GCN4 protein, a positive transcription factor in yeast, binds general control promoters at all 5' TGACTC 3' sequences. Proc Natl Acad Sci U S A. 1986 Nov;83(22):8516–8520. doi: 10.1073/pnas.83.22.8516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldari C., Cesareni G. Plasmids pEMBLY: new single-stranded shuttle vectors for the recovery and analysis of yeast DNA sequences. Gene. 1985;35(1-2):27–32. doi: 10.1016/0378-1119(85)90154-4. [DOI] [PubMed] [Google Scholar]
- Baroni M., Livian S., Martegani E., Alberghina L. Molecular cloning and regulation of the expression of the MET2 gene of Saccharomyces cerevisiae. Gene. 1986;46(1):71–78. doi: 10.1016/0378-1119(86)90168-x. [DOI] [PubMed] [Google Scholar]
- Bennetzen J. L., Hall B. D. Codon selection in yeast. J Biol Chem. 1982 Mar 25;257(6):3026–3031. [PubMed] [Google Scholar]
- Botstein D., Falco S. C., Stewart S. E., Brennan M., Scherer S., Stinchcomb D. T., Struhl K., Davis R. W. Sterile host yeasts (SHY): a eukaryotic system of biological containment for recombinant DNA experiments. Gene. 1979 Dec;8(1):17–24. doi: 10.1016/0378-1119(79)90004-0. [DOI] [PubMed] [Google Scholar]
- Bredt D. S., Hwang P. M., Glatt C. E., Lowenstein C., Reed R. R., Snyder S. H. Cloned and expressed nitric oxide synthase structurally resembles cytochrome P-450 reductase. Nature. 1991 Jun 27;351(6329):714–718. doi: 10.1038/351714a0. [DOI] [PubMed] [Google Scholar]
- Cannon M., Cannon F., Buchanan-Wollaston V., Ally D., Ally A., Beynon J. The nucleotide sequence of the nifJ gene of Klebsiella pneumoniae. Nucleic Acids Res. 1988 Dec 9;16(23):11379–11379. doi: 10.1093/nar/16.23.11379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherest H., Nguyen N. T., Surdin-Kerjan Y. Transcriptional regulation of the MET3 gene of Saccharomyces cerevisiae. Gene. 1985;34(2-3):269–281. doi: 10.1016/0378-1119(85)90136-2. [DOI] [PubMed] [Google Scholar]
- Cherest H., Surdin-Kerjan Y. Genetic analysis of a new mutation conferring cysteine auxotrophy in Saccharomyces cerevisiae: updating of the sulfur metabolism pathway. Genetics. 1992 Jan;130(1):51–58. doi: 10.1093/genetics/130.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherest H., Thomas D., Surdin-Kerjan Y. Nucleotide sequence of the MET8 gene of Saccharomyces cerevisiae. Nucleic Acids Res. 1990 Feb 11;18(3):659–659. doi: 10.1093/nar/18.3.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman B. S., Roth J. R. Genetic structure and regulation of the cysG gene in Salmonella typhimurium. J Bacteriol. 1993 Mar;175(5):1457–1466. doi: 10.1128/jb.175.5.1457-1466.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen J., Kielland-Brandt M. C. Saccharomyces carlsbergensis contains two functional MET2 alleles similar to homologues from S. cerevisiae and S. monacensis. Gene. 1994 Mar 11;140(1):33–40. doi: 10.1016/0378-1119(94)90727-7. [DOI] [PubMed] [Google Scholar]
- Henikoff S. Unidirectional digestion with exonuclease III creates targeted breakpoints for DNA sequencing. Gene. 1984 Jun;28(3):351–359. doi: 10.1016/0378-1119(84)90153-7. [DOI] [PubMed] [Google Scholar]
- Hieter P., Pridmore D., Hegemann J. H., Thomas M., Davis R. W., Philippsen P. Functional selection and analysis of yeast centromeric DNA. Cell. 1985 Oct;42(3):913–921. doi: 10.1016/0092-8674(85)90287-9. [DOI] [PubMed] [Google Scholar]
- Hoffman C. S., Winston F. A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene. 1987;57(2-3):267–272. doi: 10.1016/0378-1119(87)90131-4. [DOI] [PubMed] [Google Scholar]
- Jackson R. H., Cornish-Bowden A., Cole J. A. Prosthetic groups of the NADH-dependent nitrite reductase from Escherichia coli K12. Biochem J. 1981 Mar 1;193(3):861–867. doi: 10.1042/bj1930861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karplus P. A., Daniels M. J., Herriott J. R. Atomic structure of ferredoxin-NADP+ reductase: prototype for a structurally novel flavoenzyme family. Science. 1991 Jan 4;251(4989):60–66. [PubMed] [Google Scholar]
- Karplus P. A., Walsh K. A., Herriott J. R. Amino acid sequence of spinach ferredoxin:NADP+ oxidoreductase. Biochemistry. 1984 Dec 18;23(26):6576–6583. doi: 10.1021/bi00321a046. [DOI] [PubMed] [Google Scholar]
- Kobayashi K., Yoshimoto A. Studies on yeast sulfite reductase. IV. Structure and steady-state kinetics. Biochim Biophys Acta. 1982 Aug 10;705(3):348–356. doi: 10.1016/0167-4838(82)90257-6. [DOI] [PubMed] [Google Scholar]
- Kobayashi K., Yoshimoto A. Studies on yeast sulfite reductase. V. Effects of ionic strength on enzyme activities. Biochim Biophys Acta. 1982 Dec 6;709(1):38–45. doi: 10.1016/0167-4838(82)90418-6. [DOI] [PubMed] [Google Scholar]
- Kobayashi K., Yoshimoto A. Studies on yeast sulfite reductase. VI. Use of the effects of ionic strength as a probe for enzyme structure and mechanism. Biochim Biophys Acta. 1982 Dec 6;709(1):46–52. doi: 10.1016/0167-4838(82)90419-8. [DOI] [PubMed] [Google Scholar]
- Korch C., Mountain H. A., Byström A. S. Cloning, nucleotide sequence, and regulation of MET14, the gene encoding the APS kinase of Saccharomyces cerevisiae. Mol Gen Genet. 1991 Sep;229(1):96–108. doi: 10.1007/BF00264218. [DOI] [PubMed] [Google Scholar]
- Langin T., Faugeron G., Goyon C., Nicolas A., Rossignol J. L. The MET2 gene of Saccharomyces cerevisiae: molecular cloning and nucleotide sequence. Gene. 1986;49(3):283–293. doi: 10.1016/0378-1119(86)90364-1. [DOI] [PubMed] [Google Scholar]
- Masselot M., De Robichon-Szulmajster H. Methionine biosynthesis in Saccharomyces cerevisiae. I. Genetical analysis of auxotrophic mutants. Mol Gen Genet. 1975 Aug 5;139(2):121–132. doi: 10.1007/BF00264692. [DOI] [PubMed] [Google Scholar]
- Masselot M., Surdin-Kerjan Y. Methionine biosynthesis in Saccharomyces cerevisiae. II. Gene-enzyme relationships in the sulfate assimilation pathway. Mol Gen Genet. 1977 Jul 7;154(1):23–30. doi: 10.1007/BF00265572. [DOI] [PubMed] [Google Scholar]
- Mountain H. A., Byström A. S., Larsen J. T., Korch C. Four major transcriptional responses in the methionine/threonine biosynthetic pathway of Saccharomyces cerevisiae. Yeast. 1991 Nov;7(8):781–803. doi: 10.1002/yea.320070804. [DOI] [PubMed] [Google Scholar]
- Murphy M. J., Siegel L. M., Kamin H., Rosenthal D. Reduced nicotinamide adenine dinucleotide phosphate-sulfite reductase of enterobacteria. II. Identification of a new class of heme prosthetic group: an iron-tetrahydroporphyrin (isobacteriochlorin type) with eight carboxylic acid groups. J Biol Chem. 1973 Apr 25;248(8):2801–2814. [PubMed] [Google Scholar]
- Murphy M. J., Siegel L. M. Siroheme and sirohydrochlorin. The basis for a new type of porphyrin-related prosthetic group common to both assimilatory and dissimilatory sulfite reductases. J Biol Chem. 1973 Oct 10;248(19):6911–6919. [PubMed] [Google Scholar]
- Ostrowski J., Barber M. J., Rueger D. C., Miller B. E., Siegel L. M., Kredich N. M. Characterization of the flavoprotein moieties of NADPH-sulfite reductase from Salmonella typhimurium and Escherichia coli. Physicochemical and catalytic properties, amino acid sequence deduced from DNA sequence of cysJ, and comparison with NADPH-cytochrome P-450 reductase. J Biol Chem. 1989 Sep 25;264(27):15796–15808. [PubMed] [Google Scholar]
- Ostrowski J., Wu J. Y., Rueger D. C., Miller B. E., Siegel L. M., Kredich N. M. Characterization of the cysJIH regions of Salmonella typhimurium and Escherichia coli B. DNA sequences of cysI and cysH and a model for the siroheme-Fe4S4 active center of sulfite reductase hemoprotein based on amino acid homology with spinach nitrite reductase. J Biol Chem. 1989 Sep 15;264(26):15726–15737. [PubMed] [Google Scholar]
- Porter T. D., Kasper C. B. NADPH-cytochrome P-450 oxidoreductase: flavin mononucleotide and flavin adenine dinucleotide domains evolved from different flavoproteins. Biochemistry. 1986 Apr 8;25(7):1682–1687. doi: 10.1021/bi00355a036. [DOI] [PubMed] [Google Scholar]
- Rikkerink E. H., Magee B. B., Magee P. T. Opaque-white phenotype transition: a programmed morphological transition in Candida albicans. J Bacteriol. 1988 Feb;170(2):895–899. doi: 10.1128/jb.170.2.895-899.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sangsoda S., Cherest H., Surdin-Kerjan Y. The expression of the MET25 gene of Saccharomyces cerevisiae is regulated transcriptionally. Mol Gen Genet. 1985;200(3):407–414. doi: 10.1007/BF00425724. [DOI] [PubMed] [Google Scholar]
- Scherer S., Davis R. W. Replacement of chromosome segments with altered DNA sequences constructed in vitro. Proc Natl Acad Sci U S A. 1979 Oct;76(10):4951–4955. doi: 10.1073/pnas.76.10.4951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiestl R. H., Gietz R. D. High efficiency transformation of intact yeast cells using single stranded nucleic acids as a carrier. Curr Genet. 1989 Dec;16(5-6):339–346. doi: 10.1007/BF00340712. [DOI] [PubMed] [Google Scholar]
- Seki Y., Sogawa N., Ishimoto M. Siroheme as an active catalyst in sulfite reduction. J Biochem. 1981 Nov;90(5):1487–1492. doi: 10.1093/oxfordjournals.jbchem.a133615. [DOI] [PubMed] [Google Scholar]
- Shah V. K., Stacey G., Brill W. J. Electron transport to nitrogenase. Purification and characterization of pyruvate:flavodoxin oxidoreductase. The nifJ gene product. J Biol Chem. 1983 Oct 10;258(19):12064–12068. [PubMed] [Google Scholar]
- Siegel L. M., Davis P. S. Reduced nicotinamide adenine dinucleotide phosphate-sulfite reductase of enterobacteria. IV. The Escherichia coli hemoflavoprotein: subunit structure and dissociation into hemoprotein and flavoprotein components. J Biol Chem. 1974 Mar 10;249(5):1587–1598. [PubMed] [Google Scholar]
- Sikorski R. S., Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989 May;122(1):19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas D., Barbey R., Henry D., Surdin-Kerjan Y. Physiological analysis of mutants of Saccharomyces cerevisiae impaired in sulphate assimilation. J Gen Microbiol. 1992 Oct;138(10):2021–2028. doi: 10.1099/00221287-138-10-2021. [DOI] [PubMed] [Google Scholar]
- Thomas D., Barbey R., Surdin-Kerjan Y. Gene-enzyme relationship in the sulfate assimilation pathway of Saccharomyces cerevisiae. Study of the 3'-phosphoadenylylsulfate reductase structural gene. J Biol Chem. 1990 Sep 15;265(26):15518–15524. [PubMed] [Google Scholar]
- Thomas D., Cherest H., Surdin-Kerjan Y. Elements involved in S-adenosylmethionine-mediated regulation of the Saccharomyces cerevisiae MET25 gene. Mol Cell Biol. 1989 Aug;9(8):3292–3298. doi: 10.1128/mcb.9.8.3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas D., Jacquemin I., Surdin-Kerjan Y. MET4, a leucine zipper protein, and centromere-binding factor 1 are both required for transcriptional activation of sulfur metabolism in Saccharomyces cerevisiae. Mol Cell Biol. 1992 Apr;12(4):1719–1727. doi: 10.1128/mcb.12.4.1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas D., Rothstein R., Rosenberg N., Surdin-Kerjan Y. SAM2 encodes the second methionine S-adenosyl transferase in Saccharomyces cerevisiae: physiology and regulation of both enzymes. Mol Cell Biol. 1988 Dec;8(12):5132–5139. doi: 10.1128/mcb.8.12.5132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren M. J., Roessner C. A., Santander P. J., Scott A. I. The Escherichia coli cysG gene encodes S-adenosylmethionine-dependent uroporphyrinogen III methylase. Biochem J. 1990 Feb 1;265(3):725–729. doi: 10.1042/bj2650725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watenpaugh K. D., Sieker L. C., Jensen L. H. The binding of riboflavin-5'-phosphate in a flavoprotein: flavodoxin at 2.0-Angstrom resolution. Proc Natl Acad Sci U S A. 1973 Dec;70(12):3857–3860. doi: 10.1073/pnas.70.12.3857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yabusaki Y., Murakami H., Ohkawa H. Primary structure of Saccharomyces cerevisiae NADPH-cytochrome P450 reductase deduced from nucleotide sequence of its cloned gene. J Biochem. 1988 Jun;103(6):1004–1010. doi: 10.1093/oxfordjournals.jbchem.a122370. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- Yoshimoto A., Sato R. Studies on yeast sulfite reductase. I. Purification and characterization. Biochim Biophys Acta. 1968 Apr 2;153(3):555–575. doi: 10.1016/0005-2728(68)90185-0. [DOI] [PubMed] [Google Scholar]
- Yoshimoto A., Sato R. Studies on yeast sulfite reductase. II. Partial purification and properties of genetically incomplete sulfite reductases. Biochim Biophys Acta. 1968 Apr 2;153(3):576–588. doi: 10.1016/0005-2728(68)90186-2. [DOI] [PubMed] [Google Scholar]