Abstract
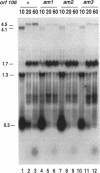
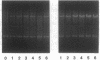
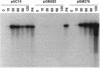
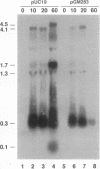
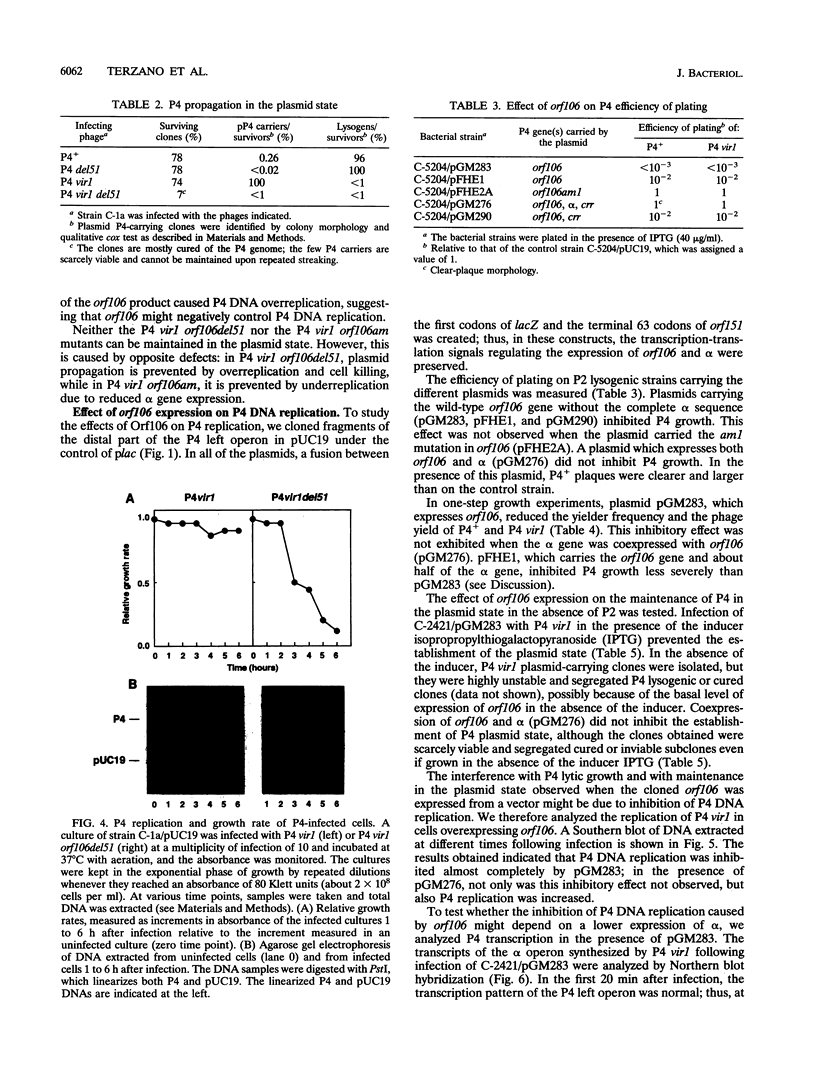
Bacteriophage P4 replication may result in either a lytic cycle or plasmid maintenance, depending on the presence or absence, respectively, of helper phase P2 genome. Bacteriophage P4 DNA replication depends on the product of gene alpha, which has origin recognition, primase, and helicase activities. An open reading frame with the coding capacity for a protein of 106 amino acids (orf106) is located upstream of the alpha gene. Genes orf106 and alpha are transcriptionally coregulated. Three amber mutations and an internal deletion (del51) were introduced into orf106. All of the amber mutations exhibited a polar effect on transcription of the downstream alpha gene. The P4 del51 mutant was slightly defective in lytic growth and could not be propagated in the plasmid state. In this latter condition, P4 DNA overreplication was observed. Overexpression of Orf106 severely inhibited P4 DNA replication, preventing P4 lytic growth and plasmid maintenance. The inhibitory effect of Orf106 on P4 replication was not observed when both orf106 and alpha were overexpressed. We suggest that orf106 is involved in P4 replication and that a balanced expression of orf106 relative to alpha may be necessary for proper P4 DNA replication. In particular, orf106 appears to be essential for the control of P4 genome replication in the plasmid state. We propose that orf106 be named cnr, for copy number regulation.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alano P., Dehò G., Sironi G., Zangrossi S. Regulation of the plasmid state of the genetic element P4. Mol Gen Genet. 1986 Jun;203(3):445–450. doi: 10.1007/BF00422069. [DOI] [PubMed] [Google Scholar]
- Barrett K. J., Marsh M. L., Calendar R. Interactions between a satellite bacteriophage and its helper. J Mol Biol. 1976 Sep 25;106(3):683–707. doi: 10.1016/0022-2836(76)90259-x. [DOI] [PubMed] [Google Scholar]
- Bertani G., Ljungquist E., Jagusztyn-Krynicka K., Jupp S. Defective particle assembly in wild type P2 bacteriophage and its correction by the lg mutation. J Gen Virol. 1978 Feb;38(2):251–261. doi: 10.1099/0022-1317-38-2-251. [DOI] [PubMed] [Google Scholar]
- Bolivar F., Rodriguez R. L., Greene P. J., Betlach M. C., Heyneker H. L., Boyer H. W., Crosa J. H., Falkow S. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2(2):95–113. [PubMed] [Google Scholar]
- Dehó G., Zangrossi S., Ghisotti D., Sironi G. Alternative promoters in the development of bacteriophage plasmid P4. J Virol. 1988 May;62(5):1697–1704. doi: 10.1128/jvi.62.5.1697-1704.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehó G., Zangrossi S., Sabbattini P., Sironi G., Ghisotti D. Bacteriophage P4 immunity controlled by small RNAs via transcription termination. Mol Microbiol. 1992 Nov;6(22):3415–3425. doi: 10.1111/j.1365-2958.1992.tb02209.x. [DOI] [PubMed] [Google Scholar]
- Flensburg J., Calendar R. Bacteriophage P4 DNA replication. Nucleotide sequence of the P4 replication gene and the cis replication region. J Mol Biol. 1987 May 20;195(2):439–445. doi: 10.1016/0022-2836(87)90664-4. [DOI] [PubMed] [Google Scholar]
- Ghisotti D., Chiaramonte R., Forti F., Zangrossi S., Sironi G., Dehò G. Genetic analysis of the immunity region of phage-plasmid P4. Mol Microbiol. 1992 Nov;6(22):3405–3413. doi: 10.1111/j.1365-2958.1992.tb02208.x. [DOI] [PubMed] [Google Scholar]
- Ghisotti D., Finkel S., Halling C., Dehò G., Sironi G., Calendar R. Nonessential region of bacteriophage P4: DNA sequence, transcription, gene products, and functions. J Virol. 1990 Jan;64(1):24–36. doi: 10.1128/jvi.64.1.24-36.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halling C., Calendar R., Christie G. E., Dale E. C., Dehò G., Finkel S., Flensburg J., Ghisotti D., Kahn M. L., Lane K. B. DNA sequence of satellite bacteriophage P4. Nucleic Acids Res. 1990 Mar 25;18(6):1649–1649. doi: 10.1093/nar/18.6.1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krevolin M. D., Inman R. B., Roof D., Kahn M., Calendar R. Bacteriophage P4 DNA replication. Location of the P4 origin. J Mol Biol. 1985 Apr 20;182(4):519–527. doi: 10.1016/0022-2836(85)90238-4. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci U S A. 1985 Jan;82(2):488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linderoth N. A., Calendar R. L. The Psu protein of bacteriophage P4 is an antitermination factor for rho-dependent transcription termination. J Bacteriol. 1991 Nov;173(21):6722–6731. doi: 10.1128/jb.173.21.6722-6731.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindqvist B. H., Dehò G., Calendar R. Mechanisms of genome propagation and helper exploitation by satellite phage P4. Microbiol Rev. 1993 Sep;57(3):683–702. doi: 10.1128/mr.57.3.683-702.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindqvist B. H., Six E. W. Replication of bacteriophage P4 DNA in a nonlysogenic host. Virology. 1971 Jan;43(1):1–7. doi: 10.1016/0042-6822(71)90218-2. [DOI] [PubMed] [Google Scholar]
- Ljungquist E., Bukhari A. I. State of prophage Mu DNA upon induction. Proc Natl Acad Sci U S A. 1977 Aug;74(8):3143–3147. doi: 10.1073/pnas.74.8.3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki I., Bertani G. Growth abnormalities in Hfr derivatives of Escherichia coli strain C. J Gen Microbiol. 1965 Sep;40(3):365–376. doi: 10.1099/00221287-40-3-365. [DOI] [PubMed] [Google Scholar]
- Schweizer H., Boos W. Transfer of the delta (argF-lac)U169 mutation between Escherichia coli strains by selection for a closely linked Tn10 insertion. Mol Gen Genet. 1983;192(1-2):293–294. doi: 10.1007/BF00327683. [DOI] [PubMed] [Google Scholar]
- Six E. W., Klug C. A. Bacteriophage P4: a satellite virus depending on a helper such as prophage P2. Virology. 1973 Feb;51(2):327–344. doi: 10.1016/0042-6822(73)90432-7. [DOI] [PubMed] [Google Scholar]
- Strack B., Lessl M., Calendar R., Lanka E. A common sequence motif, -E-G-Y-A-T-A-, identified within the primase domains of plasmid-encoded I- and P-type DNA primases and the alpha protein of the Escherichia coli satellite phage P4. J Biol Chem. 1992 Jun 25;267(18):13062–13072. [PubMed] [Google Scholar]
- Van Bokkelen G. B., Dale E. C., Halling C., Calendar R. Mutational analysis of a bacteriophage P4 late promoter. J Bacteriol. 1991 Jan;173(1):37–45. doi: 10.1128/jb.173.1.37-45.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira J., Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982 Oct;19(3):259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- Ziegelin G., Scherzinger E., Lurz R., Lanka E. Phage P4 alpha protein is multifunctional with origin recognition, helicase and primase activities. EMBO J. 1993 Sep;12(9):3703–3708. doi: 10.1002/j.1460-2075.1993.tb06045.x. [DOI] [PMC free article] [PubMed] [Google Scholar]