Gene expression in the mitochondria of kinetoplastid protists like the human pathogen Trypanosoma brucei occurs via a remarkable pathway involving both the deletion of encoded uridines (Us) and the insertion of extra Us at defined positions within mRNAs synthesized from “cryptogenes” in the maxicircle DNA (see refs. 1 and 2 for recent reviews). In some cases, >50% of the nucleotides present in the mature mRNA are generated by RNA editing events. U addition and removal create sense from nonsense, producing ORFs that encode the proteins involved in oxidative phosphorylation and other mitochondrial functions. Many of these changes are developmentally regulated, occurring only in the procyclic (insect) stage or the bloodstream (mammalian) form. Although the overall mechanism by which this extraordinary phenomenon occurs has been sketched out (3–6), filling in the details has been difficult because of the low abundance and complexity of the editing apparatus, the low efficiency of in vitro editing assays, the lack of assays for processive editing, and difficulties in assigning functions to specific proteins. The article by Aphasizhev et al. (7) in this issue of PNAS provides important information regarding the function of two of the key enzymes involved in this fascinating process.
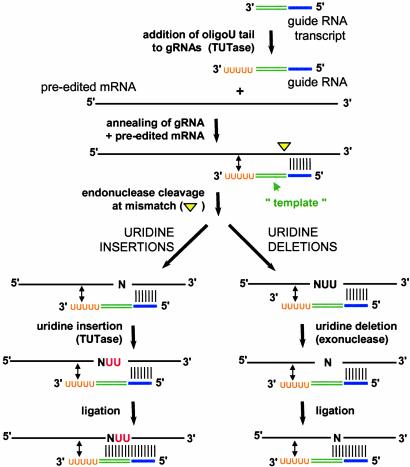
The insertion and deletion of Us into kinetoplast mRNAs occur through the concerted action of a series of enzymes (Fig. 1) (see references in ref. 2). Specificity is provided by small RNA molecules termed guide RNAs (gRNAs), which base-pair to preedited mRNAs just downstream of editing sites (8). gRNAs have three functional domains: an anchor region, which anneals to the substrate; a guiding region, which directs the insertion or deletion of U residues; and an oligo(U) tail, which is added posttranscriptionally and is thought to help tether the purine-rich 5′ cleavage fragment to the rest of the complex. Binding of the gRNA targets cleavage of the mRNA immediately upstream of the anchor duplex by an editing endonuclease, creating a 5′ cleavage fragment that is the substrate for either a 3′ terminal uridylyl transferase (TUTase) in the case of U insertion or a 3′ to 5′ exonuclease for U deletions. The number of Us added or deleted is determined by both the sequence of the gRNA and the specificity of the editing enzymes; the new 3′ end of the upstream fragment pairs with the guiding region, extending the anchor duplex and directing ligation of the two mRNA fragments. Editing of a given message occurs with an overall 3′ to 5′ polarity and usually involves multiple gRNAs that act sequentially. Often gRNA binding sites are created by prior editing events and are presumably accessed upon dissociation of the previous gRNA.
Fig. 1.
Mechanism of U insertion and deletion editing in trypanosome mitochondria. Enzymes that carry out each of the steps in U insertion and U deletion editing in trypanosome mitochondria are shown. The site of preedited mRNA cleavage upstream of the anchor duplex is shown by a yellow triangle. Added Us are shown in red. Guide RNA domains are indicated, with the anchor region in blue, the guiding region in green, and the oligo(U) tail in orange. Both A and G residues within the guiding region are used to direct U insertion. See text for details.
Editing is catalyzed by a multisubunit complex, termed the “editosome.” The size of this macromolecular complex varies, depending on the isolation conditions, but it generally sediments at 19–25S in glycerol gradients and contains between 7 and 21 polypeptides (9–13), only some of which have been directly linked to the editing reaction (see refs. 1 and 2). A number of editosome components have now been identified through sequencing and/or mass spectrometric analysis of proteins present in native complexes, and their genes have been cloned based on sequences present in the Leishmania major and T. brucei genome databases or via PCR using degenerate primers (13–16). This information has made it possible to augment conventional biochemical fractionation through purification of complexes containing individual tandem affinity purification-tagged proteins (15, 16), and the availability of antibodies raised against native complexes or recombinant proteins has facilitated affinity purification, immunodepletion, and coimmunoprecipitation studies (13, 17, 18). In addition to the enzymatic activities required for editing, a number of other proteins are present in fractions active in RNA editing or have been implicated in the editing process. These include helicases, which may be required for gRNA annealing and/or dissociation, and potential structural proteins, RNA binding proteins, and regulatory factors (see refs. 1 and 2).
In some cases it has been possible to confirm the activities of these proteins by expressing cloned genes in vitro (14, 18–20). Not surprisingly, however, it has been difficult to ascribe particular functions to the majority of the editosome components because many of these recombinant proteins do not demonstrate enzymatic activity. This could be caused by either technical issues (folding, instability, lack of posttranslational modifications, etc.) or the fact that these proteins normally function in the context of a large, multisubunit complex. Thus, genetic studies have been essential for dissecting functional roles of these components. Conventional gene knockouts and knockdowns, conditional expression or expression of dominant negative proteins in transgenic trypanosomes, and RNA interference all have been used for this purpose (7, 14, 18, 19, 21–24).
The latter approach is used in the study by Aphasizhev et al. (7) to define the roles of the two TUTases that have been identified in T. brucei RNA editing, TUTases 1 and 2 (RET1 and RET2). Previous work demonstrated that reducing the level of RET1 protein resulted in a decrease in edited mRNAs and inhibited growth (18), leading to the hypothesis that RET1 was the TUTase responsible for the addition of Us during editing. The discovery of a second TUTase in editing complexes in Leishmania tarantolae (15) and T. brucei (20) led Simpson and colleagues (7) to reexamine TUTase function, comparing the effects of down-regulation of RET1 (TbMP108) and RET2 (TbMP57) on T. brucei growth, editing complex formation, in vivo editing, in vitro editing assays, and gRNA length. These conditional RNA interference experiments demonstrated that down-regulation of RET2 inhibits growth and in vitro U insertion (but not U deletion), but has no effect on the length of gRNA tails, whereas depletion of RET1 has a more minor effect on in vitro U insertion but results in shorter gRNAs. These results implicate RET2 as the editing TUTase and RET1 as the enzyme responsible for adding oligo(U) tails to gRNAs. This work also indicates that although the U tails on gRNAs are not essential for editing in vitro (25) they are likely to be required for editing in vivo. Both recombinant proteins have TUTase activity (7, 18, 20), and the new data are consistent with previous findings that ≈40% of cellular gRNAs coimmunoprecipitate with RET1 (18) and that RET1 is not present in purified editosomes (20). Thus, it is now clear that there are two TUTases in trypanosome mitochondria and that each plays a distinct role in RNA editing.
Trypanosomes have two terminal uridylyl transferases, each playing a distinct role in RNA editing.
RET1 and RET2 are present in separate complexes (7, 20). There are also two RNA ligases implicated in editing, REL1 (TbMP52/band IV) and REL2 (TbMP48/band V), and it has been suggested that REL1 is required for deletion editing, whereas REL2 is required for U insertion (21, 26). However, other studies suggest that the functions of REL1 and REL2 may not be mutually exclusive (19, 22, 24). In addition, sub-complexes containing a subset of editosome proteins have been observed, particularly upon depletion of individual editosome components (7, 9, 15, 17, 22, 23, 27), raising a number of interesting questions regarding the mechanics of trypanosome editing. How is the switch between insertion and deletion editing effected, given that insertion and deletion sites are interspersed and often directed by the same gRNA? Are there separate complexes for insertion and deletion, or does editing occur within a single complex in which the substrate flips back and forth between insertion and deletion sites? How is the substrate shuttled among the endonuclease, TUTase or exonuclease, and ligase active sites? What roles might be played by other components identified in these complexes? How is editing regulated during development? The answers to these questions will await further dissection and eventual reconstitution of functional editing complexes from purified components.
Acknowledgments
I thank David McPheeters for critical reading of the manuscript and the National Institutes of Health for support (Grant GM054663).
See companion article on page 10617.
References
- 1.Stuart, K., Panigrahi, A. K., Schnaufer, A., Drozdz, M., Clayton, C. & Salavati, R. (2002) Philos. Trans. R. Soc. London B 357, 71–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Simpson, L., Sbicego, S. & Aphasizhev, R. (2003) RNA 9, 265–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Seiwert, S. D. & Stuart, K. (1994) Science 266, 114–117. [DOI] [PubMed] [Google Scholar]
- 4.Kable, M. L., Seiwert, S. D., Heidmann, S. & Stuart, K. (1996) Science 273, 1189–1195. [DOI] [PubMed] [Google Scholar]
- 5.Byrne, E. M., Connell, G. J. & Simpson, L. (1996) EMBO J. 15, 6758–6765. [PMC free article] [PubMed] [Google Scholar]
- 6.Cruz-Reyes, J. & Sollner-Webb, B. (1996) Proc. Natl. Acad. Sci. USA 93, 8901–8906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aphasizhev, R., Aphasizheva, I. & Simpson, L. (2003) Proc. Natl. Acad. Sci. USA 100, 10617–10622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blum, B., Bakalara, N. & Simpson, L. (1990) Cell 60, 189–198. [DOI] [PubMed] [Google Scholar]
- 9.Pollard, V. W., Harris, M. E. & Hajduk, S. L. (1992) EMBO J. 11, 4429–4438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Corell, R. A., Read, L. K., Riley, G. R., Nellissery, J. K., Allen, T. E., Kable, M. L., Wachal, M. D., Seiwert, S. D., Myler, P. J. & Stuart, K. D. (1996) Mol. Cell. Biol. 16, 1410–1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rusche, L. N., Cruz-Reyes, J., Piller, K. J. & Sollner-Webb, B. (1997) EMBO J. 16, 4069–4081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Peris, M., Simpson, A. M., Grunstein, J., Liliental, J. E., Frech, G. C. & Simpson, L. (1997) Mol. Biochem. Parasitol. 85, 9–24. [DOI] [PubMed] [Google Scholar]
- 13.Panigrahi, A. K., Gygi, S. P., Ernst, N. L., Igo, R. P., Jr., Palazzo, S. S., Schnaufer, A., Weston, D. S., Carmean, N., Salavati, R., Aebersold, R. & Stuart, K. D. (2001) Mol. Cell. Biol. 21, 380–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rusche, L. N., Huang, C. E., Piller, K. J., Hemann, M., Wirtz, E. & Sollner-Webb, B. (2001) Mol. Cell. Biol. 21, 979–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aphasizhev, R., Aphasizheva, I., Nelson, R. E., Gao, G., Simpson, A. M., Kang, X., Falick, A. M., Sbicego, S. & Simpson, L. (2003) EMBO J. 22, 913–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Panigrahi, A. K., Schnaufer, A., Ernst, N. L., Wang, B., Carmean, N., Salavati, R. & Stuart, K. (2003) RNA 9, 484–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang, C. E., O'Hearn, S. F. & Sollner-Webb, B. (2002) Mol. Cell. Biol. 22, 3194–3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aphasizhev, R., Sbicego, S., Peris, M., Jang, S. H., Aphasizheva, I., Simpson, A. M., Rivlin, A. & Simpson, L. (2002) Cell 108, 637–648. [DOI] [PubMed] [Google Scholar]
- 19.Schnaufer, A., Panigrahi, A. K., Panicucci, B., Igo, R. P., Jr., Wirtz, E., Salavati, R. & Stuart, K. (2001) Science 291, 2159–2162. [DOI] [PubMed] [Google Scholar]
- 20.Ernst, N. L., Panicucci, B., Igo, R. P., Jr., Panigrahi, A. K., Salavati, R. & Stuart, K. (2003) Mol. Cell 11, 1525–1536. [DOI] [PubMed] [Google Scholar]
- 21.Huang, C. E., Cruz-Reyes, J., Zhelonkina, A. G., O'Hearn, S., Wirtz, E. & Sollner-Webb, B. (2001) EMBO J. 20, 4694–4703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Drozdz, M., Palazzo, S. S., Salavati, R., O'Rear, J., Clayton, C. & Stuart, K. (2002) EMBO J. 21, 1791–1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang, B., Ernst, N. L., Palazzo, S. S., Panigrahi, A. K., Salavati, R. & Stuart, K. (2003) Eukaryotic Cell 2, 578–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gao, G. & Simpson, L. (2003) J. Biol. Chem. 278, 27570–27574. [DOI] [PubMed] [Google Scholar]
- 25.Burgess, M. L., Heidmann, S. & Stuart, K. (1999) RNA 5, 883–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cruz-Reyes, J., Zhelonkina, A. G., Huang, C. E. & Sollner-Webb, B. (2002) Mol. Cell. Biol. 22, 4652–4660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Madison-Antenucci, S., Sabatini, R. S., Pollard, V. W. & Hajduk, S. L. (1998) EMBO J. 17, 6368–6376. [DOI] [PMC free article] [PubMed] [Google Scholar]