Abstract
We propose that retroviruses exploit a cell-encoded pathway of intercellular vesicle traffic, exosome exchange, for both the biogenesis of retroviral particles and a low-efficiency but mechanistically important mode of infection. This Trojan exosome hypothesis reconciles current paradigms of retrovirus-directed transmission with the unique lipid composition of retroviral particles, the host cell proteins present in retroviral particles, the complex cell biology of retroviral release, and the ability of retroviruses to infect cells independently of Envelope protein–receptor interactions. An exosomal origin also predicts that retroviruses pose an unsolvable paradox for adaptive immune responses, that retroviral antigen vaccines are unlikely to provide prophylactic protection, and that alloimmunity is a central component of antiretroviral immunity. Finally, the Trojan exosome hypothesis has important implications for the fight against HIV and AIDS, including how to develop new antiretroviral therapies, assess the risk of retroviral infection, and generate effective antiretroviral vaccines.
Retroviruses are enveloped positive-strand RNA viruses that replicate through a DNA intermediate inserted in the host cell genome (1). Current models of retroviral biology adhere to basic principles of virology (2), explain most empirical data on retroviruses, and assume a complete reliance on retroviral Env proteins for the binding and fusion of retroviral particles with host cells (1, 2). However, these models do not provide a mechanistic explanation for many important properties of retroviruses, including the array of host cell molecules in retroviral particles (3–5), the observation of receptor-independent and Env-independent retroviral infections (6–8), and the ability of retroviruses to thrive in the presence of otherwise healthy adaptive immune systems (1, 2).
In an effort to reconcile these observations with the main body of data on retroviral biology, we propose the Trojan exosome hypothesis. Many eukaryotic cells synthesize and release small extracellular vesicles called exosomes, which can fuse with membranes of neighboring cells to complete an intercellular vesicle trafficking pathway (9–12). The Trojan exosome hypothesis states that retroviruses use the preexisting, nonviral exosome biogenesis pathway for the formation of infectious particles, and the preexisting, nonviral pathway of exosome uptake for a receptor-independent, Env-independent mode of infection. The following presents a portion of the empirical support for this hypothesis and its major implications for the fight against HIV and AIDS.
Exosome Biogenesis and Uptake
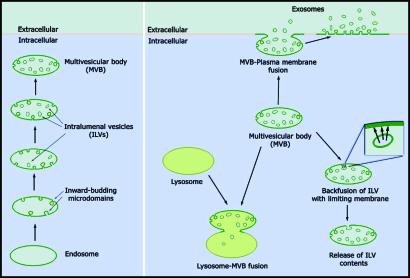
Exosomes are small (50–200 nm) membrane-bound vesicles that are released into the extracellular milieu (10–12). The early stages in exosome synthesis follow that of intralumenal vesicles (ILVs), which form by inward budding of the endosome membrane (13); endosomes enriched in ILVs are also referred to as multivesicular bodies (MVBs) (Fig. 1). The immediate fate of ILVs and their constituents can vary. For example, the ILV biogenesis pathway can be reversed, as when proteins that are targeted into discrete ILVs return to the endosomal limiting membrane (14). ILVs can also be degraded in lysosomes if the endosomes that carry them fuse with, or mature into, lysosomes (13). Alternatively, MVBs can fuse with the plasma membrane (PM), releasing ILVs into the extracellular milieu as exosomes (delayed exosome biogenesis). MVB-PM fusion also generates a patch of endosomal membrane at the cell surface that can shed exosomes directly into the extracellular fluid (immediate exosome biogenesis).
Fig. 1.
The formation (Left) and fates (Right) of ILVs.
Once released, exosomes can fuse with membranes of neighboring cells, delivering membrane and cytoplasmic proteins from one cell to another. Exosome exchange plays important roles in numerous physiological events, including prostate-induced sperm motility, wingless-mediated pattern formation, lymphocyte activation, and induction of immunological tolerance (10–12, 15, 16). Exosome uptake appears to involve clathrin-mediated endocytosis followed by backfusion of exosomes with the limiting membrane of the endosome. The net effect of the reaction is the transfer of membranes and cytosol from one cell to another in the proper topology.
Implications for Retroviral Biogenesis and Transmission
The Trojan exosome hypothesis predicts that retroviral particles and exosomes will contain a similar array of host cell lipids and proteins, use the same protein targeting and vesicle biogenesis pathway, and move between cells in the absence of a retroviral Env protein. A review of the empirical data finds support for each of these predictions.
Similarities in Host Cell Lipids and Proteins. Retroviruses and exosomes have a shared lipid composition that includes significantly higher levels of cholesterol and glycosphingolipids as compared with the plasma membrane (9, 17–19). Retroviruses and exosomes also share many protein components that are enriched relative to the PM (tetraspannins, GPI proteins and Lamps; refs. 3–5 and 20–23), membrane proteins that are present at high levels on exosomes as well as the cell surface (integrins, MHC proteins, etc.; refs. 3–5, 22, and 24–26), and numerous cytoplasmic proteins (actin, cyclophilin, tsg101, heat shock proteins, etc.; refs. 5, 22, and 27–30). Moreover, side-by-side analyses show identical host cell protein profiles for retroviral particles and exosomal preparations (31, 32). It should be noted that the host cell proteins present in retroviral particles and exosomes are not merely the abundant components of the PM (22, 23). Also, the host cell proteins in retroviruses are not just trace components, as some (MHC class II) can exceed the abundance of Env proteins (33, 34). Much of this data are from the HIV field, but similar results have been reported for other retroviruses (35–43).
Similarities in Protein Targeting and Vesicle Biogenesis. Expression of Gag alone is sufficient to drive the formation of retrovirus-like particles (44, 45). The Trojan exosome hypothesis predicts that Gag should therefore be targeted to ILVs. Protein targeting into ILVs appears to be mediated by several mechanisms, including binding to ILV components, fatty acylation, aggregation, and monoubiquitylation, and Gag proteins from several retroviruses possess these properties (13, 44). For instance, HIV Gag binds ILV components and ILV-biogenesis factors (cyclophilin, tsg101), is N-terminally myristoylated, forms large aggregates, and is monoubiquitylated (44). In addition, Gag mutants that interfere with these targeting mechanisms, such as the late domain mutants, can block the formation of retrovirus-like particles (44). Additional support for the Trojan exosome hypothesis comes from the observation that retroviral biogenesis requires ILV biogenesis factors such as tsg101 and VPS4 (44, 46).
Similarities in Cell Biology of Release. Exosomes can form at the limiting membrane of discrete endosomes (delayed exosome biogenesis) or at endosomal patches of the cell surface (immediate exosome biogenesis). The Trojan exosome hypothesis predicts that retroviruses should also form at both endosomal membranes and at patches of the cell surface, which has been observed for most, if not all retroviruses (Supporting Text, which is published as supporting information on the PNAS web site, www.pnas.org). For instance, it is firmly established that HIV buds into endosomes in macrophages and dendritic cells (47, 48). In T cells, HIV arises primarily from the cell surface (2, 8). However, HIV release in these cells is not distributed evenly across the cell surface but occurs primarily at focal patches that display late endosomal marker proteins (49, 50), which are also the sites of immediate exosome biogenesis. Interestingly, the cell surface budding of HIV in T cells is not determined by Gag, which directs HIV release, but by Vpu, as Vpu-defective strains of HIV bud into endosomes even in T cells (51–53). The Trojan exosome hypothesis is also consistent with recent evidence linking retroviral biogenesis to lipid rafts (54, 55). Like retroviruses, exosomes are enriched for cholesterol, glycosphingolipids, and many lipid raft markers, which may reflect an important role for lipid raft domains in the formation of ILVs.
Receptor-Independent and Env-Independent Infection. A defining element of the Trojan exosome hypothesis is its prediction that retroviruses will have a low-efficiency ability to infect cells independently of their Env proteins and independently of the retroviral receptors. Perhaps the clearest demonstration of Env-independent infection is that of the gypsy retrovirus, which is transmitted in vivo as efficiently in the absence of its Env gene as in the presence of its Env gene (7). Another example of Env-independent infection comes from in vitro studies of mdg3, a fly retroelement that lacks an Env-like gene even in the WT state (56).
Env-independent infection has also been documented for HIV (6, 57). The deletion of HIV Env reduces their ability to infect CD4+ cells (CD4 is the primary HIV receptor) to 1% of WT, consistent with the well established and important role of HIV Env in mediating infection of CD4+ cell types. However, removal of the Env gene does not eliminate HIV's ability to infect cells and Env-deleted HIV particles infect CD4– cells as efficiently as WT HIV particles (6, 57). An Env-independent pathway of retroviral transmission is also consistent with many other observations regarding retroviruses, such as (i) receptor-independent infection (6, 8, 58), (ii) the high-efficiency binding of Env-deleted retroviruses to cells (59–61), (iii) infection of species that lack their receptor (1, 62, 63), and (iv) the ease with which retroviral pseudotypes can be generated (1). Finally, it should be noted that the Trojan exosome hypothesis provides a greatly simplified model for the genesis of retroviruses from LTR retrotransposons (Supporting Text and Figs. 2 and 3, which are published as supporting information on the PNAS web site) as well as a model of retroviral tropism that combines both Env-dependent and Env-independent pathways (Supporting Text).
Implications for Retroviral Immunity
The Trojan exosome hypothesis is a general model of retroviral biogenesis and transmission, and thus, cannot explain the distinct properties of different retroviruses or the unique features of different retrovirus-induced pathologies. However, an exosomal origin provides retroviruses with complex physical and functional properties that have important implications for routes of pathogenesis. In the following section we will discuss how these properties allow retroviruses to thrive in the face of adaptive immune responses while rendering them susceptible to destruction by histocompatibility, or alloimmune, reactions.
The Failure of Adaptive Immunity
It is well established that humans and other mammals combat many enveloped viruses by (i) the selective proliferation of B cells that secrete neutralizing antibodies (primarily IgGs) able to block Env-mediated entry and (ii) the selective proliferation of virus-reactive T cell clones that detect and kill infected cells, amplify the antiviral immune response, and provide immunological memory (2, 64). In healthy individuals, these mechanisms are usually sufficient to either clear viruses from the body or drive viruses into latent states where replication occurs only infrequently. Furthermore, most viruses that cause acute pathogenesis (influenza, poliomyelitis, smallpox, etc.) are easily controlled in individuals with prior exposure to viral antigens. However, retroviruses are relatively resistant to adaptive responses directed at viral antigens. This is evident from the productive infection of many otherwise healthy humans by HIV (60 million infected people), human T cell leukemia virus (HTLV-1; 20 million infected people), and many other animals by many other retroviruses (1, 2, 8).
Retroviruses do not thrive by evading the adaptive immune system. In fact, most retrovirus-infected animals mount vigorous B and T cell responses to retroviral antigens (65–68). In the case of HIV, these responses (i) significantly reduce viral titers after the initial burst of viral replication early in infection (2, 8), (ii) generate antibodies that can neutralize HIV in vitro (69), and (iii) exert strong selective pressures that can shape the evolution of HIV genomes in vivo (65, 70–73). However, retroviral replication continues apace in the vast majority of HIV-infected patients. This pattern is also observed in HTLV-1-infected patients and in a variety of animals infected by their retroviruses (1, 2, 66). As for those rare HIV-infected individuals who appear to control their retroviral infection (long-term nonprogressors), it is not clear whether adaptive immune responses are responsible for this apparent control, and in some cases there is strong evidence that it is not (74–78).
Exosome Exchange and the Intrinsic Susceptibility of Immune System Cells. Retroviral resistance to adaptive immune responses is usually attributed to (i) the rapid and error-prone nature of retroviral replication, which allows for antigenic drift and the subsequent proliferation of escape mutants, and (ii) the unique problems posed by proviral insertion in host cell chromosomes (1, 2, 69). These are important factors, but the Trojan exosome hypothesis suggests the existence of several other impediments to adaptive immune control of retroviral infections.
Lymphocytes, macrophages, and immature dendritic cells commonly exchange exosomes in the process of immune surveillance and signaling (10–12, 16). Active exosome exchange among immune cells indicates that these cells will have an intrinsic, low-level susceptibility to retroviral infection via exosome exchange, in addition to whatever tropism is specified by the retroviral Env protein. This intrinsic susceptibility is enhanced by the migration of immune system cells throughout the body during immune surveillance, which exposes them to numerous exosomes from large areas of the body, and by the transmission of numerous immune cells between individuals during sex and breastfeeding (69). A major prediction from this line of argument is that retroviruses will replicate in cells of the immune system in vivo, which matches empirical observation (1, 2) (Supporting Text and Table 1, which is published as supporting information on the PNAS web site). HTLV-1 represents a particularly good example of this principle because it primarily infects CD4+ T cells in vivo even though it displays no detectable preference for these cells in vitro (2, 79).
Retroviral Targeting of Antigen-Specific T Cells. Another aspect of the Trojan exosome hypothesis that contributes to the failure of adaptive immunity is deduced from the role of exosomes in cellular immunity. Recent studies have established that exosomes are produced by nearly all antigen-presenting cells, are loaded with MHC/peptide complexes (21, 80–83), and are sufficient to stimulate and activate T cells in an MHC/peptide/T cell receptor (TCR)-dependent manner (16, 80, 84, 85). Moreover, activation of T cells stimulates their exosome exchange pathway (80, 86). Given that retrovirus-infected cells release Trojan exosomes carrying MHC proteins loaded with retroviral peptides, our hypothesis predicts the preferential infection of T cells that express retrovirus-specific TCRs. This prediction concurs with the empirical observation that HIV preferentially infects T cells that express TCRs specific for HIV antigens (87, 88). The same principle also predicts infectious synergy between retroviruses and secondary infectious agents (Supporting Text).
Retroviral Evasion of Humoral Responses. The humoral response to viruses is generally believed to function through the production of subtype G immunoglobulins (IgG) that block Env function and thereby neutralize the virus. However, an exosomal origin of retroviruses will allow them to infect neighboring cells by exosome exchange even in the presence of IgGs that completely block Env function. Empirical support for retroviral transmission in the presence of potent antibody responses comes from the observation that many productively infected AIDS patients have anti-Env antibodies that are neutralizing in vitro (67, 69). Furthermore, an exosomal origin for retroviral particles loads them with high levels of complement-inhibiting proteins (CD55, CD59; ref. 24) that protect the particles from destruction by IgG-mediated complement recruitment and lysis (26, 40, 89–93).
Ideal Immunity to Retroviral Attack
Taken together, the preceding arguments and lines of evidence indicate that retroviruses pose an unsolvable paradox for adaptive immune responses. However, the ability of retroviruses to significantly reduce host fitness, the ancient origins of retroviruses, and the ubiquity of retroviruses in the animal kingdom (1, 2) indicate that animals should possess a potent mechanism of antiretroviral immunity. The Trojan exosome hypothesis makes three predictions regarding its nature.
First, effective antiretroviral immunity in all animals must kill retroviruses and retrovirus-infected cells (i) without prior exposure to the virus and (ii) before a single round of replication has occurred in host cells. Therefore, it must be directed against exosomal antigens that are present on the surface of retroviruses and retrovirus-infected cells but are encoded by the genome of the prior host, not the retroviral genome. Second, because retroviral transmission occurs primarily within a species, the Trojan exosome hypothesis predicts that antiretroviral immunity must be directed against host cell exosomal antigens that are highly polymorphic within the population. Third, the detection of polymorphic, nonself, host cell exosomal antigens should induce a wide array of potent and naïve responses aimed at destroying nonself membranes and inhibiting retroviral replication in cells of the new host. Most animals display precisely these activities under the umbrella of their alloimmune, or histocompatibility, response. This response is shared by organisms that span the animal kingdom, from the most primitive invertebrates, the sponges, up to and including humans (94, 95). However, alloimmunity has been studied most extensively in humans, as have retroviruses, and our discussion of alloimmunity to retroviruses will rest heavily on human biology.
Evidence for the Antiretroviral Nature of Alloimmunity
Humans possess three major histocompatibility responses. The immediate and naïve response to carbohydrate alloantigens is arguably the most intense. This response requires no prior exposure to human tissue and is exemplified by the hemolytic response to incompatibility at the ABO loci. Individuals who fail to express the A or B carbohydrate antigens contain subtype M immunoglobulins (IgMs) to these antigens (because of prior exposure of B cells to these carbohydrates on commensal organisms, pathogens, etc.). These antibodies bind membranes that express the A and/or B sugars on their glycoproteins and glycoplipids and destroy these membranes by recruiting complement, including the membrane attack complex (64). The Secretor, Lewis, I, P, and T antigen loci also control the expression of polymorphic loci capable of lysing alloantigen-expressing membranes (96). The evidence that carbohydrate alloantigens are active in retroviral resistance comes from the ability of antibodies to ABO antigens to destroy retroviral particles in vitro (97) and the observation that incompatibility at the Secretor locus is associated with reduced HIV transmission in vivo (98).
The second mechanism of alloimmunity is the pleiotropic, direct alloresponse of T cells to nonself MHC protein/peptide complexes. When T cells detect alloantigenic MHC/peptide complexes, they unleash a cytotoxic attack on the alloantigen-expressing membranes (99) and release an array of soluble antiretroviral factors (100–102), even if they have never previously been exposed to the nonself MHC/peptide complex (64). Alloantigenicity at MHC loci is associated with decreased rates of HIV transmission in vivo (103–108), demonstrating that this arm of alloimmunity is an active mechanism of retroviral resistance.
The third facet of human alloimmunity is the adaptive response to histocompatibility antigens. This element of alloimmunity relies on adaptive responses to alloantigens encountered during sex, pregnancy, child-birth, breastfeeding, blood exchange, and any other form of tissue exchange. As a result, alloimmunized individuals induce both humoral and cellular responses to major and minor histocompatibility antigens, including highly polymorphic peptide blood group antigens, platelet antigens, MHC proteins, and any other antigenic polymorphisms. Evidence for alloimmunization-induced retroviral resistance is strong. Sera from multiparous women, polytransfused patients, and other alloimmunized individuals destroys HIV particles and inhibits HIV infection in vitro (108–110) and similar results have been observed in animal systems with other retroviruses (35, 36, 111). Also, vaccination studies have demonstrated that induced responses to host cell proteins present in retroviral particles can provide prophylactic resistance to retroviral infection (112–123).
Implications for Therapies, Risks, and Vaccines
HIV and HTLV-1 pose an extreme threat to human societies and individuals. The remainder of this essay discuss some aspects of the Trojan exosome hypothesis that are relevant to the development of antiretroviral therapies, the role of histocompatibility in retroviral transmission, and the design of antiretroviral vaccines.
Therapies. An exosomal origin for retroviral particles should render them sensitive to inhibitors of exosome biogenesis, and inhibitors of exosome uptake should enhance retroviral susceptibility to inhibitors of Env-dependent infection (124). Both classes of drugs might be found among the antiretroviral factors released in extracts of alloreactive cells from a wide variety of organisms. In terms of gene therapy, destructive catalytic domains (RNases, proteases, lipases, etc.) fused to cytoplasmic exosomal proteins (TSG101, cyclophilins, etc.) or to the cytoplasmic tail of exosomal membrane proteins (MHC class II, tetraspannins, etc.) might inhibit retroviral transmission by degrading molecules that are important for exosome biogenesis and/or exosome uptake. However, deleterious side effects may result from therapies that disrupt exosome exchange. A more specific gene therapy approach might involve the use of zymogens that are activated only in response to specific retroviral factors, such as the HIV and HTLV-1 proteases. Finally, the ability of IgM class antibodies to destroy retroviral particles and retrovirus-infected cells via complement activation, despite the presence of complement-inhibiting proteins on the retroviral surface, indicates that recombinant IgM molecules directed against specific retroviral epitopes might be useful in attenuating retroviral infections and perhaps eliminating some retroviral reservoirs, particularly when combined with other antiretroviral therapies.
Histocompatibility and the Risk of Infection. The probability of retroviral transmission during intimate contact between two individuals can be affected by the mode of tissue transfer and the amount of tissue transferred (type of sex, breastfeeding, transplantation, etc.) (2, 8). The Trojan exosome hypothesis predicts that the degree of histocompatibility is another important risk factor vis-à-vis retroviral transmission. More specifically, this hypothesis predicts relatively efficient retroviral transmission between individuals who happen to be histocompatible and relatively inefficient retroviral transmission between individuals who happen to be histoincompatible. Such a risk factor might help explain why HIV is transmitted through the largely outbred human population, on average, only once per ≈200 exposures even though some individuals are infected after just a single exposure (2, 8, 125). It might also contribute to the observation that the risk of HIV infection increases for people who engage in tissue exchange with multiple partners, as this would increase their odds of encountering an infected histocompatible individual (2, 8, 125).
The Design of Antiretroviral Vaccines. The Trojan exosome hypothesis and the empirical record support the concept of alloimmunity as a major mechanism of retroviral resistance. Given that alloimmunity is dramatically enhanced by prior exposure to alloantigens, alloimmunization is a logical tool to use in combating the spread of retroviruses through animal populations. Alloimmunization is currently approved as a treatment for certain types of infertility in humans, and appears to be relatively free from adverse health consequences (110, 126). Not surprisingly, two of the researchers who have demonstrated a role for alloimmunity in retroviral resistance, Shearer and Lehner, reached the conclusion that alloimmunization should be applied in the fight against HIV (110, 127, 128). The Trojan exosome hypothesis provides additional support for this approach.
For a monogamous couple, alloimmunization can be accomplished rather easily (110, 127, 128). However, the HIV and HTLV-1 pandemics are being driven by nonmonogamous sexual behaviors and other forms of promiscuous tissue exchange (2, 8, 125). Allovaccines must therefore be designed to induce prophylactic immunity against numerous different alloantigens encoded by many histocompatibility alleles that exist in a given population. One strategy is to immunize individuals with the most common alloantigens of their population. Moreover, such vaccines do not necessarily require high technology, because the most potent allovaccines might well consist of nothing more than pooled, inactivated cells from an appropriate set of donors. Another alloimmunization strategy is to exploit the potential of sex-specific antigens. Although this approach will be limited to blocking heterosexual transmission, male-to-female transmission appears to be the major route of HIV spread in many societies. Thus, immunizing females with male-derived blood cells or recombinant H–Y antigens might enhance their resistance to retroviral infections and help thwart the spread of HIV and other retroviruses. A third approach is to immunize individuals with arrays of polymeric carbohydrates that induce the production of specific IgMs and thereby maximize the effect of carbohydrate alloantigens in retroviral resistance.
An important feature of alloimmunization strategies is that they are directed against host cell-derived antigens rather than rapidly evolving retroviral antigens, and thus, are less encumbered by antigenic differences between different retroviral strains than are conventional vaccination strategies. For example, alloimmunization of humans has the potential to induce prophylactic protection to all strains of HIV as well as to HTLV-1 and any other retroviruses that may be moving through the human population. However, alloimmunization poses some unusual risks for individuals and societies that also deserve consideration (Supporting Text).
The Consequences of Viral Antigen Immunization. We previously discussed how an exosomal origin together with other facets of retroviral biology allows retroviruses to persist and thrive even in the presence of potent adaptive immune responses. For the same reasons, the Trojan exosome hypothesis predicts vaccines based on retroviral proteins are unlikely to be successful. This prediction is supported by the empirical evidence that retroviral antigen-based vaccines are unable to induce prophylactic protection in vaccinated individuals (112, 118, 125, 129–131).
A more insidious prediction of the Trojan exosome hypothesis vis-à-vis retroviral antigen vaccines is that they might potentiate subsequent infection and/or pathogenesis. The preferential infection of retroviral antigen-specific T cells (87, 88) raises the specter that vaccines that induce the proliferation of such T cells will render vaccinated individuals more susceptible to infection during subsequent exposures. In addition, such vaccines might accelerate the progression of retrovirus-induced pathogenesis by enhancing the infection of T cells that are involved in suppressing retroviral infections. This concern is not merely theoretical, because vaccine-induced pathogenesis has been observed in horses and cats immunized with recombinant retroviral Env proteins from equine infectious anemia virus and feline leukemia virus, respectively (112, 118, 129, 130). Whole killed retroviruses are composed of both alloantigens and retroviral antigens and presents a complex set of advantages and disadvantages (Supporting Text).
Conclusions
The Trojan exosome hypothesis proposes a mechanism of retroviral evolution, biogenesis, and transmission that explains the wide range of physical and functional properties of retroviruses. Although the Trojan exosome hypothesis does not explain the distinguishing pathologies caused by different retroviruses, it does offer a mechanistic basis for some of the most clinically important aspects of retroviral infection. These include the ability of retroviruses to replicate in animals with otherwise healthy immune systems, the susceptibility of retroviruses to alloimmune responses, and the ineffectiveness of viral antigen-based vaccines. These predictions have important implications for human health. HIV is already responsible for 20 million deaths, will kill 40 million people within the next decade, and infects >15,000 people each day (2, 8, 125). In addition, HTLV-1 has infected 20 million individuals and is also spreading at a rapid pace (2). Therapeutic approaches can repress HIV replication in certain infected individuals but they are unlikely to halt the spread of HIV through human populations. This goal might only be achieved through the development of prophylactic antiretroviral vaccines.
To be successful, any antiretroviral vaccination strategy must take account of the fundamental cell biology of retroviral biogenesis and transmission, the empirical data regarding the induction of prophylactic resistance to retroviral infection, and the evolutionarily relevant mechanisms of antiretroviral resistance. The Trojan exosome is a coherent cell biological model that reconciles the broad array of data on retroviral biology into a single mechanism of retroviral biogenesis and transmission. It is supported by a wide array of empirical evidence. Finally, it is an integral component of a model for evolution under selection by cell-associated pathogens that explains all relevant properties of alloimmunity in organisms as diverse as sponges and humans (unpublished observation). By these most basic criteria, alloimmunization is a vaccination strategy that should be used in the fight against HIV.
Supplementary Material
Acknowledgments
We thank J. Berg, J. Boeke, S. Desiderio, R. Doolittle, G. Hart, J. Levy, J. Nathans, P. Parham, K. Sacksteder, R. Tinker, R. Tinker-Kulberg, A. Varki, C. Wills, and members of the Gould and Hildreth laboratories for stimulating discussions and insight. We also thank J. Cohen for his enlightening book (132). Finally, we recognize the many researchers whose work we could not acknowledge here. The authors are supported by grants from the National Institutes of Health and The Johns Hopkins Fund for Medical Discovery.
References
- 1.Coffin, J. M., Hughes, S. H. & Varmus, H. E. (1997) in Retroviruses (Cold Spring Harbor Lab. Press, Plainview, NY), p. 843. [PubMed]
- 2.Knipe, D. M. & Howley, P. M. (2001) Fields Virology (Lippincott Williams & Wilkins, Philadelphia).
- 3.Arthur, L. O., Bess, J. W., Jr., Sowder, R. C., II, Benveniste, R. E., Mann, D. L., Chermann, J. C. & Henderson, L. E. (1992) Science 258, 1935–1938. [DOI] [PubMed] [Google Scholar]
- 4.Orentas, R. J. & Hildreth, J. E. (1993) AIDS Res. Hum. Retroviruses 9, 1157–1165. [DOI] [PubMed] [Google Scholar]
- 5.Ott, D. E. (1997) Rev. Med. Virol. 7, 167–180. [DOI] [PubMed] [Google Scholar]
- 6.Pang, S., Yu, D., An, D. S., Baldwin, G. C., Xie, Y., Poon, B., Chow, Y. H., Park, N. H. & Chen, I. S. (2000) J. Virol. 74, 10994–11000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chalvet, F., Teysset, L., Terzian, C., Prud'homme, N., Santamaria, P., Bucheton, A. & Pelisson, A. (1999) EMBO J. 18, 2659–2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Levy, J. A. (1998) HIV and the Pathogenesis of AIDS (Am. Soc. Microbiol., Washington, DC).
- 9.Johnstone, R. M., Adam, M., Hammond, J. R., Orr, L. & Turbide, C. (1987) J. Biol. Chem. 262, 9412–9420. [PubMed] [Google Scholar]
- 10.Denzer, K., Kleijmeer, M. J., Heijnen, H. F., Stoorvogel, W. & Geuze, H. J. (2000) J. Cell Sci. 113, 3365–3374. [DOI] [PubMed] [Google Scholar]
- 11.Stoorvogel, W., Kleijmeer, M. J., Geuze, H. J. & Raposo, G. (2002) Traffic 3, 321–330. [DOI] [PubMed] [Google Scholar]
- 12.Thery, C., Zitvogel, L. & Amigorena, S. (2002) Nat. Rev. Immunol. 2, 569–579. [DOI] [PubMed] [Google Scholar]
- 13.Katzmann, D. J., Odorizzi, G. & Emr, S. D. (2002) Nat. Rev. Mol. Cell Biol. 3, 893–905. [DOI] [PubMed] [Google Scholar]
- 14.Hirst, J., Futter, C. E. & Hopkins, C. R. (1998) Mol. Biol. Cell 9, 809–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Greco, V., Hannus, M. & Eaton, S. (2001) Cell 106, 633–645. [DOI] [PubMed] [Google Scholar]
- 16.Hwang, I., Shen, X. & Sprent, J. (2003) Proc. Natl. Acad. Sci. USA 100, 6670–6675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aloia, R. C., Jensen, F. C., Curtain, C. C., Mobley, P. W. & Gordon, L. M. (1988) Proc. Natl. Acad. Sci. USA 85, 900–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aloia, R. C., Tian, H. & Jensen, F. C. (1993) Proc. Natl. Acad. Sci. USA 90, 5181–5185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mobius, W., Ohno-Iwashita, Y., van Donselaar, E. G., Oorschot, V. M., Shimada, Y., Fujimoto, T., Heijnen, H. F., Geuze, H. J. & Slot, J. W. (2002) J. Histochem. Cytochem. 50, 43–55. [DOI] [PubMed] [Google Scholar]
- 20.Sakalian, M. & Hunter, E. (1998) Adv. Exp. Med. Biol. 440, 329–339. [DOI] [PubMed] [Google Scholar]
- 21.Escola, J. M., Kleijmeer, M. J., Stoorvogel, W., Griffith, J. M., Yoshie, O. & Geuze, H. J. (1998) J. Biol. Chem. 273, 20121–20127. [DOI] [PubMed] [Google Scholar]
- 22.Thery, C., Boussac, M., Veron, P., Ricciardi-Castagnoli, P., Raposo, G., Garin, J. & Amigorena, S. (2001) J. Immunol. 166, 7309–7318. [DOI] [PubMed] [Google Scholar]
- 23.Nguyen, D. H. & Hildreth, J. E. (2000) J. Virol. 74, 3264–3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rabesandratana, H., Toutant, J. P., Reggio, H. & Vidal, M. (1998) Blood 91, 2573–2580. [PubMed] [Google Scholar]
- 25.Esser, M. T., Graham, D. R., Coren, L. V., Trubey, C. M., Bess, J. W., Jr., Arthur, L. O., Ott, D. E. & Lifson, J. D. (2001) J. Virol. 75, 6173–6182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marschang, P., Sodroski, J., Wurzner, R. & Dierich, M. P. (1995) Eur. J. Immunol. 25, 285–290. [DOI] [PubMed] [Google Scholar]
- 27.Luban, J., Bossolt, K. L., Franke, E. K., Kalpana, G. V. & Goff, S. P. (1993) Cell 73, 1067–1078. [DOI] [PubMed] [Google Scholar]
- 28.Gurer, C., Cimarelli, A. & Luban, J. (2002) J. Virol. 76, 4666–4670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mathew, A., Bell, A. & Johnstone, R. M. (1995) Biochem. J. 308, 823–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thery, C., Regnault, A., Garin, J., Wolfers, J., Zitvogel, L., Ricciardi-Castagnoli, P., Raposo, G. & Amigorena, S. (1999) J. Cell Biol. 147, 599–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hammarstedt, M., Wallengren, K., Pedersen, K. W., Roos, N. & Garoff, H. (2000) Proc. Natl. Acad. Sci. USA 97, 7527–7532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bess, J. W., Jr., Gorelick, R. J., Bosche, W. J., Henderson, L. E. & Arthur, L. O. (1997) Virology 230, 134–144. [DOI] [PubMed] [Google Scholar]
- 33.Ott, D. E. & Henderson, L. E. (1995) AIDS Vaccine Program (Science Applications International and the National Cancer Institute, Frederick, MD), pp. III/10–III/14.
- 34.Gelderblom, H. R., Ozel, M., Winkel, T., Morath, B., Grund, C. & Pauli, G. (1991) in Accessory Cells in HIV and Other Retroviral Infections: Morphological and Functional Aspects, eds. Racz, P., Dijkstra, J. C. & Gluckman, J. C. (Karger, Basel), pp. 50–68.
- 35.Azocar, J. & Essex, M. (1979) Cancer Res. 39, 3388–3391. [PubMed] [Google Scholar]
- 36.Azocar, J., Essex, M. & Yunis, E. J. (1983) Hum. Immunol. 7, 59–65. [DOI] [PubMed] [Google Scholar]
- 37.Bubbers, J. E. & Lilly, F. (1977) Nature 266, 458–459. [DOI] [PubMed] [Google Scholar]
- 38.Hildreth, J. E., Subramanium, A. & Hampton, R. A. (1997) J. Virol. 71, 1173–1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hildreth, J. E. (1998) J. Virol. 72, 9544–9552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hiasa, A., Watanabe, M., Okada, H., Ikenaka, K., Fujita, T., Yoshimatsu, T., Kanematsu, T. & Shiku, H. (1999) Int. J. Oncol. 14, 1091–1096. [DOI] [PubMed] [Google Scholar]
- 41.Mortara, R. A. & Koch, G. L. (1989) J. Submicrosc. Cytol. Pathol. 21, 295–306. [PubMed] [Google Scholar]
- 42.Strehlow, D., Jodo, S. & Ju, S. T. (2000) Proc. Natl. Acad. Sci. USA 97, 4209–4214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jodo, S., Strehlow, D. & Ju, S. T. (2000) J. Immunol. 164, 5062–5069. [DOI] [PubMed] [Google Scholar]
- 44.Pornillos, O., Garrus, J. E. & Sundquist, W. I. (2002) Trends Cell Biol. 12, 569–579. [DOI] [PubMed] [Google Scholar]
- 45.Strack, B., Calistri, A., Accola, M. A., Palu, G. & Gottlinger, H. G. (2000) Proc. Natl. Acad. Sci. USA 97, 13063–13068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Garrus, J. E., von Schwedler, U. K., Pornillos, O. W., Morham, S. G., Zavitz, K. H., Wang, H. E., Wettstein, D. A., Stray, K. M., Cote, M., Rich, R. L., et al. (2001) Cell 107, 55–65. [DOI] [PubMed] [Google Scholar]
- 47.Blom, J., Nielsen, C. & Rhodes, J. M. (1993) APMIS 101, 672–680. [DOI] [PubMed] [Google Scholar]
- 48.Raposo, G., Moore, M., Innes, D., Leijendekker, R., Leigh-Brown, A., Benaroch, P. & Geuze, H. (2002) Traffic 3, 718–729. [DOI] [PubMed] [Google Scholar]
- 49.Leng, Q., Bentwich, Z., Magen, E., Kalinkovich, A. & Borkow, G. (2002) AIDS 16, 519–529. [DOI] [PubMed] [Google Scholar]
- 50.Miranda, L. R., Schaefer, B. C., Kupfer, A., Hu, Z. & Franzusoff, A. (2002) Proc. Natl. Acad. Sci. USA 99, 8031–8036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Klimkait, T., Strebel, K., Hoggan, M. D., Martin, M. A. & Orenstein, J. M. (1990) J. Virol. 64, 621–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gottlinger, H. G., Dorfman, T., Cohen, E. A. & Haseltine, W. A. (1993) Proc. Natl. Acad. Sci. USA 90, 7381–7385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li, Q. G., Zhang, Y. J., Liang, Y., Feng, C. Q., Li, Y. Z., Sjoberg, R., Jiang, Y., Wang, N. F. & Wadell, G. (1995) J. Acquired Immune Defic. Syndr. Hum. Retrovirol. 9, 103–113. [PubMed] [Google Scholar]
- 54.Liao, Z., Cimakasky, L. M., Hampton, R., Nguyen, D. H. & Hildreth, J. E. (2001) AIDS Res. Hum. Retroviruses 17, 1009–1019. [DOI] [PubMed] [Google Scholar]
- 55.Niyogi, K. & Hildreth, J. E. (2001) J. Virol. 75, 7351–7361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Syomin, B. V., Leonova, T. Y. & Ilyin, Y. V. (2002) Mol. Genet. Genomics 267, 418–423. [DOI] [PubMed] [Google Scholar]
- 57.Chow, Y. H., Yu, D., Zhang, J. Y., Xie, Y., Wei, O. L., Chiu, C., Foroohar, M., Yang, O. O., Park, N. H., Chen, I. S., et al. (2002) J. Acquired Immune Defic. Syndr. 30, 1–8. [DOI] [PubMed] [Google Scholar]
- 58.Marras, D., Bruggeman, L. A., Gao, F., Tanji, N., Mansukhani, M. M., Cara, A., Ross, M. D., Gusella, G. L., Benson, G., D'Agati, V. D., et al. (2002) Nat. Med. 8, 522–526. [DOI] [PubMed] [Google Scholar]
- 59.Pizzato, M., Blair, E. D., Fling, M., Kopf, J., Tomassetti, A., Weiss, R. A. & Takeuchi, Y. (2001) Gene Ther. 8, 1088–96. [DOI] [PubMed] [Google Scholar]
- 60.Pizzato, M., Marlow, S. A., Blair, E. D. & Takeuchi, Y. (1999) J. Virol. 73, 8599–8611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Walker, S. J., Pizzato, M., Takeuchi, Y. & Devereux, S. (2002) J. Virol. 76, 6909–6918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Johnston, J. B., Olson, M. E., Rud, E. W. & Power, C. (2001) Curr. Biol. 11, 1109–1113. [DOI] [PubMed] [Google Scholar]
- 63.Weiss, R. A., Magre, S. & Takeuchi, Y. (2000) J. Infect. 40, 21–25. [DOI] [PubMed] [Google Scholar]
- 64.Janeway, C. A., Travers, P., Walport, M. & Shlomik, M. (2001) Immunobiology (Garland, New York).
- 65.Rimmelzwaan, G. F., Siebelink, K. H., Broos, H., Drost, G. A., Weijer, K., van Herwijnen, R. & Osterhaus, A. D. (1994) Vet. Microbiol. 39, 153–165. [DOI] [PubMed] [Google Scholar]
- 66.Sellon, D. C. (1993) Vet. Clin. North Am. Equine Pract. 9, 321–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Connor, R. I., Korber, B. T., Graham, B. S., Hahn, B. H., Ho, D. D., Walker, B. D., Neumann, A. U., Vermund, S. H., Mestecky, J., Jackson, S., et al. (1998) J. Virol. 72, 1552–1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Addo, M. M., Yu, X. G., Rathod, A., Cohen, D., Eldridge, R. L., Strick, D., Johnston, M. N., Corcoran, C., Wurcel, A. G., Fitzpatrick, C. A., et al. (2003) J. Virol. 77, 2081–2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sabin, A. B. (1992) Proc. Natl. Acad. Sci. USA 89, 8852–8855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Burns, D. P. & Desrosiers, R. C. (1994) Curr. Top. Microbiol. Immunol. 188, 185–219. [DOI] [PubMed] [Google Scholar]
- 71.Goulder, P. J., Brander, C., Tang, Y., Tremblay, C., Colbert, R. A., Addo, M. M., Rosenberg, E. S., Nguyen, T., Allen, R., Trocha, A., et al. (2001) Nature 412, 334–338. [DOI] [PubMed] [Google Scholar]
- 72.Burns, D. P., Collignon, C. & Desrosiers, R. C. (1993) J. Virol. 67, 4104–4113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Carpenter, S., Evans, L. H., Sevoian, M. & Chesebro, B. (1987) J. Virol. 61, 3783–3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhang, L., Yu, W., He, T., Yu, J., Caffrey, R. E., Dalmasso, E. A., Fu, S., Pham, T., Mei, J., Ho, J. J., et al. (2002) Science 298, 995–1000. [DOI] [PubMed] [Google Scholar]
- 75.Altfeld, M., Allen, T. M., Yu, X. G., Johnston, M. N., Agrawal, D., Korber, B. T., Montefiori, D. C., O'Connor, D. H., Davis, B. T., Lee, P. K., et al. (2002) Nature 420, 434–439. [DOI] [PubMed] [Google Scholar]
- 76.Alexander, L., Weiskopf, E., Greenough, T. C., Gaddis, N. C., Auerbach, M. R., Malim, M. H., O'Brien, S. J., Walker, B. D., Sullivan, J. L. & Desrosiers, R. C. (2000) J. Virol. 74, 4361–4376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Alexander, L., Aquino-DeJesus, M. J., Chan, M. & Andiman, W. A. (2002) J. Virol. 76, 10533–10539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kirchhoff, F., Greenough, T. C., Brettler, D. B., Sullivan, J. L. & Desrosiers, R. C. (1995) N. Engl. J. Med. 332, 228–232. [DOI] [PubMed] [Google Scholar]
- 79.Richardson, J. H., Edwards, A. J., Cruickshank, J. K., Rudge, P. & Dalgleish, A. G. (1990) J. Virol. 64, 5682–5687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Raposo, G., Nijman, H. W., Stoorvogel, W., Liejendekker, R., Harding, C. V., Melief, C. J. & Geuze, H. J. (1996) J. Exp. Med. 183, 1161–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Clayton, A., Court, J., Navabi, H., Adams, M., Mason, M. D., Hobot, J. A., Newman, G. R. & Jasani, B. (2001) J. Immunol. Methods 247, 163–174. [DOI] [PubMed] [Google Scholar]
- 82.Denzer, K., van Eijk, M., Kleijmeer, M. J., Jakobson, E., de Groot, C. & Geuze, H. J. (2000) J. Immunol. 165, 1259–1265. [DOI] [PubMed] [Google Scholar]
- 83.Wolfers, J., Lozier, A., Raposo, G., Regnault, A., Thery, C., Masurier, C., Flament, C., Pouzieux, S., Faure, F., Tursz, T., et al. (2001) Nat. Med. 7, 297–303. [DOI] [PubMed] [Google Scholar]
- 84.Skokos, D., Le Panse, S., Villa, I., Rousselle, J. C., Peronet, R., Namane, A., David, B. & Mecheri, S. (2001) Int. Arch. Allergy Immunol. 124, 133–136. [DOI] [PubMed] [Google Scholar]
- 85.Skokos, D., Le Panse, S., Villa, I., Rousselle, J. C., Peronet, R., David, B., Namane, A. & Mecheri, S. (2001) J. Immunol. 166, 868–876. [DOI] [PubMed] [Google Scholar]
- 86.Blanchard, N., Lankar, D., Faure, F., Regnault, A., Dumont, C., Raposo, G. & Hivroz, C. (2002) J. Immunol. 168, 3235–3241. [DOI] [PubMed] [Google Scholar]
- 87.Douek, D. C., Brenchley, J. M., Betts, M. R., Ambrozak, D. R., Hill, B. J., Okamoto, Y., Casazza, J. P., Kuruppu, J., Kunstman, K., Wolinsky, S., et al. (2002) Nature 417, 95–98. [DOI] [PubMed] [Google Scholar]
- 88.Demoustier, A., Gubler, B., Lambotte, O., De Goer, M. G., Wallon, C., Goujard, C., Delfraissy, J. F. & Taoufik, Y. (2002) AIDS 16, 1749–1754. [DOI] [PubMed] [Google Scholar]
- 89.Montefiori, D. C., Cornell, R. J., Zhou, J. Y., Zhou, J. T., Hirsch, V. M. & Johnson, P. R. (1994) Virology 205, 82–92. [DOI] [PubMed] [Google Scholar]
- 90.Saifuddin, M., Ghassemi, M., Patki, C., Parker, C. J. & Spear, G. T. (1994) AIDS Res. Hum. Retroviruses 10, 829–837. [DOI] [PubMed] [Google Scholar]
- 91.Saifuddin, M., Parker, C. J., Peeples, M. E., Gorny, M. K., Zolla-Pazner, S., Ghassemi, M., Rooney, I. A., Atkinson, J. P. & Spear, G. T. (1995) J. Exp. Med. 182, 501–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Stoiber, H., Pinter, C., Siccardi, A. G., Clivio, A. & Dierich, M. P. (1996) J. Exp. Med. 183, 307–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Schmitz, J., Zimmer, J. P., Kluxen, B., Aries, S., Bogel, M., Gigli, I. & Schmitz, H. (1995) J. Clin. Invest. 96, 1520–1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bodmer, W. F. (1972) Nature 237, 139–145. [DOI] [PubMed] [Google Scholar]
- 95.Humphreys, T. & Reinherz, E. L. (1994) Immunol. Today 15, 316–320. [DOI] [PubMed] [Google Scholar]
- 96.Spitalnik, P. F. & Spitalnik, S. L. (2000) in Hematology, eds. Hoffman, R. W., Benz, E. J., Shattil, S. J., Furie, B., Cohen, H. J., Silberstein, L. E. & McGlave, P. (Churchill Livingstone, Philadelphia), pp. 2188–2196.
- 97.Arendrup, M., Hansen, J. E., Clausen, H., Nielsen, C., Mathiesen, L. R. & Nielsen, J. O. (1991) AIDS 5, 441–444. [DOI] [PubMed] [Google Scholar]
- 98.Ali, S., Niang, M. A., N'Doye, I., Critchlow, C. W., Hawes, S. E., Hill, A. V. & Kiviat, N. B. (2000) J. Infect. Dis. 181, 737–739. [DOI] [PubMed] [Google Scholar]
- 99.Hernandez-Fuentes, M. P., Baker, R. J. & Lechler, R. I. (1999) Rev. Immunogenet. 1, 282–296. [PubMed] [Google Scholar]
- 100.Walker, C. M., Moody, D. J., Stites, D. P. & Levy, J. A. (1986) Science 234, 1563–1566. [DOI] [PubMed] [Google Scholar]
- 101.Pinto, L. A., Blazevic, V., Patterson, B. K., Mac Trubey, C., Dolan, M. J. & Shearer, G. M. (2000) J. Virol. 74, 4505–4511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Pinto, L. A., Blazevic, V., Shearer, G. M., Patterson, B. K. & Dolan, M. J. (2000) Blood 95, 1875–1876. [PubMed] [Google Scholar]
- 103.Polycarpou, A., Ntais, C., Korber, B. T., Elrich, H. A., Winchester, R., Krogstad, P., Wolinsky, S., Rostron, T., Rowland-Jones, S. L., Ammann, A. J., et al. (2002) AIDS Res. Hum. Retroviruses 18, 741–746. [DOI] [PubMed] [Google Scholar]
- 104.Hader, S. L., Hodge, T. W., Buchacz, K. A., Bray, R. A., Padian, N. S., Rausa, A., Slaviniski, S. A. & Holmberg, S. D. (2002) J. Infect. Dis. 185, 1729–1735. [DOI] [PubMed] [Google Scholar]
- 105.Beyrer, C., Artenstein, A. W., Rugpao, S., Stephens, H., VanCott, T. C., Robb, M. L., Rinkaew, M., Birx, D. L., Khamboonruang, C., Zimmerman, P. A., et al. (1999) J. Infect. Dis. 179, 59–67. [DOI] [PubMed] [Google Scholar]
- 106.MacDonald, K. S., Embree, J., Njenga, S., Nagelkerke, N. J., Ngatia, I., Mohammed, Z., Barber, B. H., Ndinya-Achola, J., Bwayo, J. & Plummer, F. A. (1998) J. Infect. Dis. 177, 551–556. [DOI] [PubMed] [Google Scholar]
- 107.Wang, Y., Tao, L., Mitchell, E., Bravery, C., Berlingieri, P., Armstrong, P., Vaughan, R., Underwood, J. & Lehner, T. (1999) Nat. Med. 5, 1004–1009. [DOI] [PubMed] [Google Scholar]
- 108.Wang, Y., Underwood, J., Vaughan, R., Harmer, A., Doyle, C. & Lehner, T. (2002) Clin. Exp. Immunol. 129, 493–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Spruth, M., Stoiber, H., Kacani, L., Schonitzer, D. & Dierich, M. P. (1999) AIDS Res. Hum. Retroviruses 15, 533–543. [DOI] [PubMed] [Google Scholar]
- 110.Kiprov, D. D., Sheppard, H. W. & Hanson, C. V. (1994) Science 263, 737–738. [DOI] [PubMed] [Google Scholar]
- 111.Aupoix, M., Vigier, P. & Blanchet, J. P. (1980) J. Gen. Virol. 46, 63–73. [DOI] [PubMed] [Google Scholar]
- 112.Pedersen, N. C., Johnson, L., Birch, D. & Theilen, G. H. (1986) Vet. Immunol. Immunopathol. 11, 123–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Murphey-Corb, M., Martin, L. N., Davison-Fairburn, B., Montelaro, R. C., Miller, M., West, M., Ohkawa, S., Baskin, G. B., Zhang, J. Y., Putney, S. D., et al. (1989) Science 246, 1293–1297. [DOI] [PubMed] [Google Scholar]
- 114.Stott, E. J., Chan, W. L., Mills, K. H., Page, M., Taffs, F., Cranage, M., Greenaway, P. & Kitchin, P. (1990) Lancet 336, 1538–1541. [DOI] [PubMed] [Google Scholar]
- 115.Stott, E. J. (1991) Nature 353, 393. [DOI] [PubMed] [Google Scholar]
- 116.Langlois, A. J., Weinhold, K. J., Matthews, T. J., Greenberg, M. L. & Bolognesi, D. P. (1992) AIDS Res. Hum. Retroviruses 8, 1641–1652. [DOI] [PubMed] [Google Scholar]
- 117.Chan, W. L., Rodgers, A., Hancock, R. D., Taffs, F., Kitchin, P., Farrar, G. & Liew, F. Y. (1992) J. Exp. Med. 176, 1203–1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Issel, C. J., Horohov, D. W., Lea, D. F., Adams, W. V., Jr., Hagius, S. D., McManus, J. M., Allison, A. C. & Montelaro, R. C. (1992) J. Virol. 66, 3398–3408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Johnson, P. R., Montefiori, D. C., Goldstein, S., Hamm, T. E., Zhou, J., Kitov, S., Haigwood, N. L., Misher, L., London, W. T., Gerin, J. L., et al. (1992) Proc. Natl. Acad. Sci. USA 89, 2175–2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Johnson, P. R., Montefiori, D. C., Goldstein, S., Hamm, T. E., Zhou, J., Kitov, S., Haigwood, N. L., Misher, L., London, W. T., Gerin, J. L., et al. (1992) AIDS Res. Hum. Retroviruses 8, 1501–1505. [DOI] [PubMed] [Google Scholar]
- 121.Goldstein, S., Elkins, W. R., London, W. T., Hahn, A., Goeken, R., Martin, J. E. & Hirsch, V. M. (1994) J. Med. Primatol. 23, 75–82. [DOI] [PubMed] [Google Scholar]
- 122.Elyar, J. S., Tellier, M. C., Soos, J. M. & Yamamoto, J. K. (1997) Vaccine 15, 1437–1444. [DOI] [PubMed] [Google Scholar]
- 123.Hoover, E. A., Mullins, J. I., Chu, H. J. & Wasmoen, T. L. (1996) AIDS Res. Hum. Retroviruses 12, 379–383. [DOI] [PubMed] [Google Scholar]
- 124.Kilby, J. M., Hopkins, S., Venetta, T. M., DiMassimo, B., Cloud, G. A., Lee, J. Y., Alldredge, L., Hunter, E., Lambert, D., Bolognesi, D., et al. (1998) Nat. Med. 4, 1302–1307. [DOI] [PubMed] [Google Scholar]
- 125.Graham, B. S. (2002) Annu. Rev. Med. 53, 207–221. [DOI] [PubMed] [Google Scholar]
- 126.Kiprov, D. D., Nachtigall, R. D., Weaver, R. C., Jacobson, A., Main, E. K. & Garovoy, M. R. (1996) Am. J. Reprod. Immunol. 36, 228–234. [DOI] [PubMed] [Google Scholar]
- 127.Lehner, T., Shearer, G. M., Hackett, C. J., Schultz, A. & Sharma, O. K. (2000) AIDS Res. Hum. Retroviruses 16, 309–313. [DOI] [PubMed] [Google Scholar]
- 128.Shearer, G. M., Pinto, L. A. & Clerici, M. (1999) Immunol. Today 20, 66–71. [DOI] [PubMed] [Google Scholar]
- 129.Siebelink, K. H., Tijhaar, E., Huisman, R. C., Huisman, W., de Ronde, A., Darby, I. H., Francis, M. J., Rimmelzwaan, G. F. & Osterhaus, A. D. (1995) J. Virol. 69, 3704–3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Wang, S. Z., Rushlow, K. E., Issel, C. J., Cook, R. F., Cook, S. J., Raabe, M. L., Chong, Y. H., Costa, L. & Montelaro, R. C. (1994) Virology 199, 247–251. [DOI] [PubMed] [Google Scholar]
- 131.Feinberg, M. B. & Moore, J. P. (2002) Nat. Med. 8, 207–210. [DOI] [PubMed] [Google Scholar]
- 132.Cohen, J. (2001) Shots in the Dark: The Wayward Search for an AIDS Vaccine (W. W. Norton, New York).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.