Abstract
For HIV-1 to enter a cell, its envelope protein (Env) must sequentially engage CD4 and a chemokine coreceptor, triggering conformational changes in Env that ultimately lead to fusion between the viral and host cell membranes. Each step of the virus entry pathway is a potential target for novel antiviral agents termed entry inhibitors. A growing number of entry inhibitors are under clinical development, with one having already been licensed by the Food and Drug Administration. With the emergence of virus strains that are largely resistant to existing reverse transcriptase and protease inhibitors, the development of entry inhibitors comes at an opportune time. Nonetheless, because all entry inhibitors target in some manner the highly variable Env protein of HIV-1, there are likely to be challenges in their efficient application that are unique to this class of drugs. Env density, receptor expression levels, and differences in affinity and receptor presentation are all factors that could influence the clinical response to this promising class of new antiviral agents.
Anew class of anti-HIV-1 drugs has been developed: compounds known variously as fusion or entry inhibitors (1, 2). The most clinically advanced entry inhibitor, T20 (known now as enfuvirtide) from Trimeris (Durham, NC), has now been licensed by the Food and Drug Administration. Many other compounds are presently in or will soon approach earlier-stage clinical trials. Clinical efficacy in the sense of drug-induced reductions in plasma viremia has been shown for several entry inhibitors including those that block membrane fusion (3–5), binding of the viral gp120 protein to the CD4 receptor (6), and binding of gp120 to either the CCR5 (B. Baroudy and M. Laughlin, personal communication) or CXCR4 (G. Bridger, personal communication) coreceptors. Hence, it seems likely that entry inhibitors will prove to be effective additions to the reverse-transcriptase (RT) and protease inhibitors that are presently used to treat HIV-1 infection. It can be anticipated, however, that entry inhibitors will need to be used in combination with these other antiretrovirals for long-term suppression of circulating virus to be achieved. It is also likely that resistance to entry inhibitors will arise and that viral genotyping and phenotyping will probably become important clinical tests that will help guide entry-inhibitor therapy. In addition, there are several issues relating to the safety and application of entry inhibitors that are predictable enough from preclinical and early clinical data to warrant discussion here.
Mechanism of HIV-1 Entry and Its Inhibition
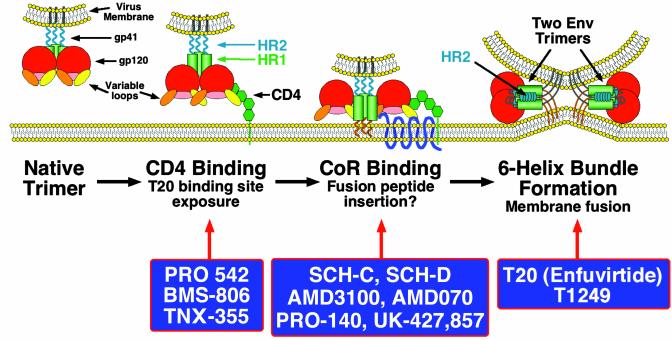
The development of entry inhibitors has been facilitated by the discovery of the cellular receptors needed for virus infection and by the consequent understanding of the receptor-induced conformational changes in the viral envelope (Env) protein that lead to virus-cell fusion (7–9). Env is a homotrimeric type I integral membrane protein; each Env subunit consists of a gp120 surface protein that mediates binding to cellular receptors and a noncovalently associated gp41 transmembrane protein that has a hydrophobic fusion peptide at its N terminus (7). For HIV-1 to enter a cell, Env must be triggered to undergo conformational changes that mediate fusion between the viral and cellular membranes (Fig. 1). The first step in the fusion process entails binding of gp120 subunits to cell surface CD4 molecules. The structure of a large portion of gp120 in complex with CD4 has been determined, revealing a conserved “pocket” into which a region of CD4 inserts (10). PRO 542, a tetrameric, CD4-based chimeric protein consisting of four gp120-binding domains fused to IgG2 Fc regions, can neutralize primary viruses by preventing CD4 binding (11). In addition, the conserved CD4-binding pocket on gp120 is a target for BMS-806, a small molecule with potent antiviral activity against primary isolates in vitro (12).
Fig. 1.
A model for HIV entry is shown, with the steps prevented by different entry inhibitors shown rather than the step at which each entry inhibitor binds. For example, T20 binds to Env after it engages CD4 (second section), but it blocks six-helix bundle formation (fourth section). BMS-806 binds to the native Env (first section) and prevents binding to CD4 (second section).The Env protein is a homotrimeric protein with each subunit containing surface gp120 and membrane-spanning gp41 proteins. The native Env trimer is a target for neutralizing antibodies (first section), although few other broadly cross-reactive, neutralizing antibodies have been described. Binding to CD4 is mediated by the gp120 subunit, and this can be inhibited by the small molecule inhibitor BMS-806 (Bristol-Myers Squibb) by the CD4-IgG2 chimera PRO 542 (Progenics, Tarrytown, NY) and by the anti-CD4 antibody TNX-355 (Tanox, Houston) (second section). CD4 binding induces conformational changes in gp120 that result in the exposure of a conserved region that participates in coreceptor binding (second section). This conserved region is, in the native trimer, hidden in part by variable loops that are thought to be repositioned after CD4 binding. Although only a single CD4-binding event is shown, multiple CD4-binding events may be needed to activate a single Env trimer. CD4 binding also makes Env a target for the fusion inhibitor T20, a peptide that binds to a triple-stranded coiled coil in the N-terminal region of gp41 that is formed by the helical HR1 domains (shown by green cylinders). A more potent fusion inhibitor that has completed early clinical trials, T1249, also targets this region. It is not known with what efficiency the T20-binding site is exposed by CD4 binding alone, and it is possible that coreceptor binding may be needed to cause full exposure. After CD4 binding, gp120 binds to a seven-transmembrane domain coreceptor (third section, CoR). Coreceptor binding can be inhibited by several CCR5 blockers that are under clinical development including SCH-C and SCH-D (both from Schering-Plough) and UK-427,857 (from Pfizer, Sandwich, U.K.; C. Hitchcock, personal communication), by the anti-CCR5 antibody PRO-140 (Progenics), and by the CXCR4 inhibitors AMD3100 and AMD070 (both from AnorMED, Vancouver). The hydrophobic fusion peptide at the N terminus of gp41 becomes exposed and inserts into the membrane of the cell. Whether this results from CD4 binding or coreceptor binding is not known. Coreceptor binding ultimately results in formation of a six-helix bundle in which the helical HR2 domains in each gp41 subunit fold back and pack into grooves on the outside of the triple-stranded HR1 domains (fourth section), bringing the fusion peptide and transmembrane domain of gp41 (and their associated membranes) into close proximity. It is likely that several Env trimers need to undergo this conformational change in order to form a fusion pore, although here only two trimers are depicted. It is not known whether gp120 remains associated during the fusion process or dissociates from gp41.
Although CD4 binding is required for infection by the vast majority of primary HIV-1 strains, it is not sufficient by itself. A coreceptor is also necessary, usually one of the chemokine receptors CCR5 or CXCR4 (7–9). Coreceptor binding is made possible by the conformational changes induced in gp120 by CD4 binding; the resulting structural rearrangements of gp120 domains create or expose the coreceptor-binding site (13). Together, CD4 and coreceptor binding induce additional conformational changes in gp41, including exposure of the fusion peptide, which is first displaced toward the cell membrane and then inserts into it (8). These processes seem to be mediated in part by the formation of a triple-stranded coiled coil from the N-terminal helical regions, termed HR1, of each of the three gp41 ectodomains (Fig. 1). Formation of this structure can be induced by CD4 binding alone (14–16). The gp41 subunit then folds back on itself, allowing a second, more C-terminal helical region, termed HR2, to pack into grooves on the outside of the triple-stranded coiled coil. Eventually, a six-helix bundle is formed, comprising three HR1 domains in the center, with three HR2 domains packed on the outside in an antiparallel fashion (Fig. 1). As a result of this transition, the fusion peptide and transmembrane domain of gp41, along with their associated membranes, are brought into close proximity (17, 18). The change in free energy associated with this structural transition is predicted to be sufficient to cause lipid mixing and membrane fusion (8, 14).
T20 is a peptide based on the sequence of the HR2 region. It binds to the triple-stranded coiled coil formed by the three HR1 domains, thereby preventing formation of the six-helix bundle and hence inhibiting membrane fusion (19). Thus, T20 targets a structural intermediate of the fusion process: It does not bind to native Env, nor does it bind to the six-helix bundle. Rather, it binds to a transient Env conformation that is induced by CD4 binding (14–16) but lost when the six-helix bundle forms.
Virus-Dependent Influences on the Efficacy of Entry Inhibitors
The complexity of the virus-cell fusion process outlined above allows for several different, to some extent interlocking, influences on the potency with which different inhibitors can antagonize entry. Because all entry inhibitors target the viral Env protein directly (e.g., PRO 542 or T20) or indirectly (e.g., coreceptor blockers) and Env is the most variable of the HIV-1 proteins, it is not surprising that primary virus strains differ in their sensitivity to entry inhibitors. Moreover, the extent of strain-to-strain variation is markedly greater for entry inhibitors than it is for RT and protease inhibitors (20). The actual viral factors that influence entry-inhibitor sensitivity are only now being identified and understood, but they presumably involve variation in both gp120 and gp41.
As a general principle, it can be anticipated that the more rapid and efficient the rate of fusion, the less active any entry inhibitor is likely to be (21). Hence, viral factors that increase fusion efficiency will probably act to reduce the potency of all entry inhibitors, perhaps even including neutralizing antibodies. Although Env is the principal viral determinant of entry efficiency, a subsidiary influence, at least in theory, could be the p17 “matrix” product of the gag gene, which plays an important role in Env incorporation into virions. Theoretically, changes in p17 could affect the density of Env on the virion surface, which could increase viral resistance against any inhibitors directed at Env, including neutralizing antibodies (22). It is unknown whether natural or inhibitor-selected differences in Env incorporation caused by changes in Env or Gag sequences will be relevant in practice, however.
A more important viral influence on entry-inhibitor action is likely to be conferred by sequence variation that affects Env function directly. Env is an exceptionally plastic protein in that it can tolerate an enormous degree of sequence variation with seemingly minimal effects on how well it works (23). In general, the higher the affinity of virion-associated Env for CD4 and coreceptor, the less vulnerable the receptor-binding stages of entry will be to inhibition. In fact, enhanced affinity for coreceptors resulting from changes in either the V3 loop or the bridging sheet region of gp120 correlates with increased fusion kinetics, and thence resistance to both T20 and coreceptor inhibitors (21). Changes in Env away from the receptor binding sites could also influence entry-inhibitor sensitivity. As an example, the accessibility of mAb epitopes (and presumably also the receptor-binding sites) within the gp120 components of the Env trimer can be affected by single amino acid changes in the extracellular domain of gp41 (24) and even by sequence variation within the gp41 cytoplasmic domain (25). The extensive glycosylation of Env is another variable that could influence how efficiently mAbs bind to gp120 (26) and even, perhaps, the activity of gp41-binding compounds such as T20. A yet further complication is that the Env complex must retain its natural resistance to the binding of neutralizing antibodies in vivo even under the selection pressure of entry inhibitors; a virus that becomes neutralization-sensitive will not persist. HIV-1 and its animal counterparts probably pay a price for the structural devices that protect them from the humoral immune system by reducing their affinity for receptors and hence their rate of fusion (11). In vitro, the defenses perhaps can be weakened, because the virus trades protection for fusion efficiency (27). Overall, sequence variation almost anywhere in Env could affect its fusion function, the vulnerability of the entry process to various inhibitors, and the way HIV-1 escapes from them.
Host-Dependent Influences on the Efficacy of Entry Inhibitors
That virally encoded factors affect the potency of an anti-HIV-1 drug would scarcely be news to anyone who has studied RT and protease inhibitors. However, host factors also come into play with entry inhibitors in a way that simply does not apply for the older classes of drugs. Indeed, it is likely that infected individuals will respond quite differently to entry inhibitors. There are several reasons to believe this will be so, and the limited clinical experience gained from the phase I trials of SCH-C in humans tends to confirm the supposition. Thus, viral load declines after 10 days of dosing with an intermediate dose of SCH-C varied from 0 to >1.5 log in 12 different individuals infected with only R5 viruses (B. Baroudy and M. Laughlin, personal communication). An obvious variable is the inherent sensitivity of the infecting strain to the effect of a CCR5 inhibitor, which can differ significantly from strain to strain in vitro (28, 29). However, all the viruses isolated from the individuals in the clinical cohort discussed above had an approximately equal sensitivity to SCH-C in vitro (B. Baroudy, personal communication). The observed variation therefore may have an alternative explanation. Some of it could be a pharmacological effect relating to differences in drug absorption and metabolism between individuals, but another possibility lies in the dynamics of coreceptor-dependent HIV-1 entry. Specifically, the rate of virus-cell fusion is influenced by the density of CCR5 coreceptors on the cell surface (21), with higher levels of coreceptor being linked to enhanced fusion kinetics and increased resistance to T20 and coreceptor inhibitors (21). Moreover, the potency of a CCR5 inhibitor as a competitive inhibitor of virus-cell fusion will be inversely proportional to the density of CCR5 receptors available on the cell surface. In humans, baseline CCR5 expression on freshly isolated peripheral blood mononuclear cells spans at least a 20-fold range among humans who possess two wild-type coding alleles (30–32). Clearly, this degree of variation is significantly greater than the 2-fold reduction caused by possession of an allele encoding the defective CCR5-Δ32 protein, a gene-dosing effect sufficient to reduce the rate of disease progression (30). It can be presumed, although it has never been demonstrated properly, that a major influence on CCR5 expression in vivo is variation in the upstream regulatory regions of CCR5. Several common polymorphisms in these regions have been linked to increased or reduced rates of disease progression in HIV-1-infected people, probably by altering the extent of CCR5 protein production in one or more tissues (33).
An additional influence on CCR5 expression is immune activation caused by intercurrent infections, which may be of particular importance for understanding the pathogenesis of HIV-1 infection in regions of Africa where parasitic infections are relatively common (34). Minor coding polymorphisms in CCR5 have also been described that could possibly have an impact on the efficacy of CCR5 inhibitors but at too low of a population frequency («1%) for them to have much relevance in clinical practice (35). A far greater but as yet almost completely undefined influence could be any cellular factors that affect the rate of CCR5 recycling either basally or in response to the binding of a small molecule inhibitor. In other words, any cellular parameter that significantly alters the expression of free, functional CCR5 on the target cell surface either naturally or in response to the binding of a CCR5 inhibitor could have a profound influence on the efficacy of an entry inhibitor, particularly a CCR5 inhibitor. Finally, any differences in CCR5 processing (such as sulfation) that changed the conformation or surface availability of the coreceptor could also affect sensitivity to entry inhibitors (36).
The functional expression of CCR5, and hence the vulnerability of CCR5-mediated HIV-1 entry to antagonism by an exogenous inhibitor, also could be modulated by CC-chemokine secretion. The natural chemokine ligands of CCR5 include MIP-1α, MIP-1β, and regulated on activation normal T cell expressed and secreted (RANTES), molecules that act to inhibit HIV-1 entry in vitro by reducing access of the virus to its coreceptor and/or by causing coreceptor down-regulation (9). The expression of these CC-chemokines affects disease progression rates in HIV-1-infected people; the higher the levels of RANTES expression in peripheral blood mononuclear cells ex vivo, the slower the individual progresses to AIDS and death (37). Genetic influences on CC-chemokine expression have been documented and linked to rates of disease progression, such as a polymorphism in the RANTES promoter (33). In addition, the less CCR5 that is expressed on CD4+ T cells, at least in vitro, the more CC-chemokines are secreted because of the existence of a regulatory loop that modulates the levels of both the receptor and its ligands (30). Hence, a reduction of CCR5 expression does not just reduce the efficiency of HIV-1 entry; it can also increase the local concentration of inhibitors of CCR5-mediated entry, creating a nonlinear relationship between CCR5 expression and HIV-1 replication.
How variation in CC-chemokine levels affects the antiviral activity of a small molecule inhibitor of CCR5 is therefore complex because of the existence of several interconnecting variables. These include competition between the CC-chemokine and the small molecule inhibitor for CCR5 binding (and also for HIV-1 gp120 binding), variation in the extent to which the binding sites on CCR5 for the different ligands (CC-chemokine, gp120, and small molecule) physically overlap, and the different ways in which the various ligands might cause short- and long-term down-regulation of CCR5. Time is another variable. Some of the effects discussed above may be of short duration, such as the physical competition between a CCR5 inhibitor and an HIV-1 virion for an overlapping binding site on CCR5. However, other events could conceivably last longer, for example, CCR5 down-regulation, CC-chemokine up-regulation, or any presence of inhibitor-blocked CCR5 on the cell surface that is sustained even in the absence of free inhibitor. Much remains to be learned from clinical trials or experiments in animal models. Many of the issues discussed above apply, of course, also to CXCR4 inhibitors.
Resistance Pathways and Safety Considerations
It is to be expected that HIV-1 will evolve resistance to entry inhibitors just as it does to the existing classes of antiviral drugs (38), which is likely to necessitate the development of new clinical tests. The evolution of resistance will be minimized, but of course not eliminated, by the use of entry inhibitors in combination either with each other or existing RT and protease inhibitors. It will be important both to determine how the resistance of HIV-1 to any given entry inhibitor influences its sensitivity to other entry inhibitors and to understand the relationship between resistance and viral pathogenesis.
Thus far, there is very little information available on the development of resistance to entry inhibitors either in vitro or in vivo. T20 resistance has been studied in vitro and is now being documented in vivo (39). Maximal resistance to T20 in vitro seems to involve the generation of single amino acid changes in the region of gp41 to which T20 binds (40). Similar mutations have also been observed in vivo, particularly in patients who receive suboptimal doses of T20 (41). Resistance to T1249, a more potent analog of T20, has not been described yet in vitro or in vivo, although it will surely occur. Changes elsewhere in Env, including the V3 loop and the coreceptor-binding site in gp120, may affect fusion kinetics and can also influence sensitivity to T20 and T1249 (21).
Resistance to coreceptor inhibitors can occur via two different mechanisms. A virus can either acquire changes in Env that enable it to engage the coreceptor differently, whether or not the inhibitor is present, or it can switch the coreceptor it uses, for example from CCR5 to CXCR4. Because the presence of viruses that use CXCR4 in vivo is associated with a poor prognosis, the latter escape pathway is a point of concern, particularly if CXCR4 use persisted after the cessation of CCR5 inhibitor therapy. There is little or no evidence to suggest that coreceptors other than CCR5 and CXCR4 are important for HIV-1 infection in vivo. To date, most studies designed to select for viral resistance to coreceptor inhibitors used cell lines that express only a single coreceptor. Thus, coreceptor switching was not possible, and drug resistance was invariably associated with alterations in how the virus bound to its original coreceptor (42–47). Whether these changes created a different binding site for the coreceptor that was not affected by the inhibitor or resulted in enhanced coreceptor affinity has not been determined. Enhanced coreceptor affinity itself might result in increased pathogenicity by enabling HIV-1 to infect cells that express only low levels of coreceptor (48). Finally, the development of high-level resistance to coreceptor inhibitors in vitro has invariably involved multiple sequence changes in gp120 (45, 47, 49). Were this to be the norm in vivo, evolution of resistant viruses would be neither simple nor rapid.
Coreceptor switching is possible when HIV-1 is grown in the presence of a coreceptor inhibitor on cells that express both CCR5 and CXCR4, although it does not always occur (49, 50). When an uncloned, R5 primary isolate was cultured in human peripheral blood mononuclear cells with increasing concentrations of the small molecule CCR5 inhibitor, AD101, the virus evolved to use CCR5 in an AD101-insensitive manner (49). Both the selection of resistant variants from an initial quasispecies and de novo mutation were involved in AD101-resistance development (S. E. Kuhmann and J.P.M., unpublished results). Similar results have been obtained with a second R5 virus and with a different CCR5 inhibitor, SCH-C (ref. 49 and J. Strizki and B. Baroudy, personal communication), and with other small molecule CCR5 inhibitors (S. E. Kuhmann and J.P.M., unpublished results). In contrast, when the CXCR4 inhibitor AMD3100 was applied to peripheral blood mononuclear cells infected with X4 or R5X4 viruses, R5 viruses expanded within the cultures, reflecting either de novo evolution of R5 escape mutants or the selection and expansion of variants able to use CCR5 (51, 52). Thus, the few completed in vitro studies indicate that CCR5 is the preferred coreceptor at least under the experimental conditions used thus far. Whether this will always be the case or whether the particular pathway followed will be a stochastic process even in vitro will require additional studies with different input viruses and different CCR5 inhibitors. Similar caveats apply, in principle, to the clinical use of CXCR4 inhibitors. Of course, the possibility of coreceptor switching would be reduced by the simultaneous use of CCR5 and CXCR4 inhibitors, if both classes of drug could be developed.
Implications for Clinical Monitoring and Treatment
At present, CD4 counts and virus-load measurements are the major quantitative tests that are used to determine when to treat patients and when to change treatment regimens. Viral genotyping, in which virus is analyzed for specific mutations known to impart resistance to RT and protease inhibitors, is becoming increasingly common (53, 54). We anticipate that the use of entry inhibitors will have implications for clinical monitoring. It will be important to assess the relative proportions of R5, R5X4, and X4 viruses in individuals scheduled to receive coreceptor blockers, because CCR5 inhibitors are unlikely to significantly benefit patients harboring mostly X4 viruses, and vice versa for CXCR4 inhibitors. In addition, the possible evolution of X4 viruses in individuals receiving CCR5 inhibitors will have to be monitored particularly carefully, because their emergence would be an indication to stop or change therapy.
In addition to viral phenotyping, both viral and host genotypic tests might be developed to predict the probability of treatment success or even to adjust drug dosing. It is possible, for example, that Δ32-ccr5 or CCR5 promoter genotyping could prove useful because of their actual or potential effect on CCR5 expression levels, which in turn influence the potency of both fusion inhibitors and coreceptor inhibitors (21). In some cases, for inhibitors that actually interact with Env directly, it might be possible to make some assessments of how sequence variation affects the binding of the compound. For example, the binding site for T20 on gp41 is known in broad outline, and the principal variation that occurs under drug-selection pressure involves amino acid changes in this region (40), although other changes in Env are also likely to influence T20 sensitivity (21). Clearly, greater understanding of both viral and host factors that affect the potency of entry inhibitors would facilitate the development of new clinical monitoring tools that could make the application of entry inhibitors more efficient, increase the probability of treatment success, and limit the evolution of drug-resistant variants.
Conclusions
We believe that entry inhibitors will be important additions to the weapons now used to combat HIV-1 infection. They may also have an equally valuable role to play as topical microbicides or even systemically applied prophylactic drugs to prevent the sexual transmission of HIV-1, because in the absence of an effective vaccine, some form of biological intervention to reduce the spread of this virus is a public health priority. Likewise, the use of entry inhibitors to prevent maternal–infant transmission should also be considered. Although there is likely to be variation in how different individuals respond to the same inhibitor over and above what is conferred by virus-dependent factors, most recipients are likely to benefit substantially from entry inhibitors. This applies particularly to people who have failed therapy with conventional drugs or who will do so in the future, but treatment-naive patients should also respond extremely well. Viral escape from entry inhibitors is to be expected, and the process may be more complex (and hence arguably slower) than clinicians have been used to with protease and RT inhibitors. Issues surrounding the possible evolution of HIV-1 to a more virulent form will need to be monitored carefully, particularly with CCR5 inhibitors. Indeed, screening assays to avoid the use of these drugs in patients for whom they are unsuitable may have to be used routinely. Overall, the application of different types of entry inhibitor in combination with each other and with existing drugs is an obvious step to take for good reasons that have emerged from preclinical studies (55).
References
- 1.Moore, J. P. & Stevenson, M. (2000) Nat. Rev. Mol. Cell Biol. 1, 40–49. [DOI] [PubMed] [Google Scholar]
- 2.O'Hara, B. M. & Olson, W. C. (2002) Curr. Opin. Pharmacol. 2, 523–528. [DOI] [PubMed] [Google Scholar]
- 3.Kilby, J. M., Hopkins, S., Venetta, T. M., DiMassimo, B., Cloud, G. A., Lee, J. Y., Alldredge, L., Hunter, E., Lambert, D., Bolognesi, D., et al. (1998) Nat. Med. 4, 1302–1307. [DOI] [PubMed] [Google Scholar]
- 4.Kilby, J. M., Lalezari, J. P., Eron, J. J., Carlson, M., Cohen, C., Arduino, R. C., Goodgame, J. C., Gallant, J. E., Volberding, P., Murphy, R. L., et al. (2002) AIDS Res. Hum. Retroviruses 18, 685–693. [DOI] [PubMed] [Google Scholar]
- 5.Lalezari, J. P., Henry, K., O'Hearn, M., Montaner, J. S. G., Piliero, P. J., Trottier, B., Walmsley, S., Cohen, C., Kuritzkes, D. R., Eron, J. J., et al. (2003) N. Engl. J. Med. 348, 2175–2185. [DOI] [PubMed] [Google Scholar]
- 6.Jacobson, J. M., Lowy, I., Fletcher, C. V., O'Neill, T. J., Tran, D. N., Ketas, T. J., Trkola, A., Klotman, M. E., Maddon, P. J., Olson, W. C. & Israel, R. J. (2000) J. Infect. Dis. 182, 326–329. [DOI] [PubMed] [Google Scholar]
- 7.Wyatt, R. & Sodroski, J. (1998) Science 280, 1884–1888. [DOI] [PubMed] [Google Scholar]
- 8.Doms, R. W. & Moore, J. P. (2000) J. Cell Biol. 151, F9–F14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berger, E. A., Murphy, P. M. & Farber, J. M. (1999) Annu. Rev. Immunol. 17, 657–700. [DOI] [PubMed] [Google Scholar]
- 10.Kwong, P. D., Wyatt, R., Robinson, J., Sweet, R. W., Sodroski, J. & Hendrickson, W. A. (1998) Nature 393, 648–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Parren, P. W., Moore, J. P., Burton, D. R. & Sattentau, Q. J. (1999) AIDS 13, S137–S162. [PubMed] [Google Scholar]
- 12.Lin, P.-F., Blair, W., Wang, T., Spicer, T., Guo, Q., Zhou, N., Gong, Y.-F., Wang, H.-G. H., Rose, R., Yamanaka, G., et al. (2003) Proc. Natl. Acad. Sci. USA 100, 11013–11018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rizzuto, C. D., Wyatt, R., Hernandez-Ramos, N., Sun, Y., Kwong, P. D., Hendrickson, W. A. & Sodroski, J. (1998) Science 280, 1949–1953. [DOI] [PubMed] [Google Scholar]
- 14.Melikyan, G. B., Markosyan, R. M., Hemmati, H., Delmedico, M. K., Lambert, D. M. & Cohen, F. S. (2000) J. Cell Biol. 151, 413–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.He, Y., Vassell, R., Zaitseva, M., Nguyen, N., Yang, Z., Weng, Y. & Weiss, C. D. (2003) J. Virol. 77, 1666–1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gallo, S. A., Puri, A. & Blumenthal, R. (2001) Biochemistry 40, 12231–12236. [DOI] [PubMed] [Google Scholar]
- 17.Chan, D. C., Fass, D., Berger, J. M. & Kim, P. S. (1997) Cell 89, 263–273. [DOI] [PubMed] [Google Scholar]
- 18.Weissenhorn, W., Dessen, A., Harrison, S. C., Skehel, J. J. & Wiley, D. C. (1997) Nature 387, 426–430. [DOI] [PubMed] [Google Scholar]
- 19.Chen, C. H., Matthews, T. J., McDanal, C. B., Bolognesi, D. P. & Greenberg, M. L. (1995) J. Virol. 69, 3771–3777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Labrosse, B., Labernardière, J.-L., Dam, E., Trouplin, V., Skrabal, K., Clavel, F. & Mammano, F. (2003) J. Virol. 77, 1610–1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reeves, J. D., Gallo, S. A., Ahmad, N., Miamidian, J. L., Harvey, P. E., Sharron, M., Pöhlmann, S., Sfakianos, J. N., Derdeyn, C. A., Blumenthal, R., et al. (2002) Proc. Natl. Acad. Sci. USA 99, 16249–16254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klasse, P. J. & Moore, J. P. (1996) J. Virol. 70, 3668–3677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gaschen, B., Taylor, J., Yusim, K., Foley, B., Gao, F., Lang, D., Novitsky, V., Haynes, B., Hahn, B. H., Bhattacharya, T. & Korber, B. (2002) Science 296, 2354–2360. [DOI] [PubMed] [Google Scholar]
- 24.Klasse, P. J., McKeating, J. A., Schutten, M., Reitz, M. S., Jr., & Robert-Guroff, M. (1993) Virology 196, 332–337. [DOI] [PubMed] [Google Scholar]
- 25.Edwards, T. G., Wyss, S., Reeves, J. D., Zolla-Pazner, S., Hoxie, J. A., Doms, R. W. & Baribaud, F. (2002) J. Virol. 76, 2683–2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Olofsson, S. & Hansen, J. E. (1998) Scand. J. Infect. Dis. 30, 435–440. [DOI] [PubMed] [Google Scholar]
- 27.Moore, J. P. & Ho, D. D. (1995) AIDS 9, Suppl. A, S117–S136. [PubMed] [Google Scholar]
- 28.Baba, M., Nishimura, O., Kanzaki, N., Okamoto, M., Sawada, H., Iizawa, Y., Shiraishi, M., Aramaki, Y., Okonogi, K., Ogawa, Y., et al. (1999) Proc. Natl. Acad. Sci. USA 96, 5698–5703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Strizki, J. M., Xu, S., Wagner, N. E., Wojcik, L., Liu, J., Hou, Y., Endres, M., Palani, A., Shapiro, S., Clader, J. W., et al. (2001) Proc. Natl. Acad. Sci. USA 98, 12718–12723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu, L., Paxton, W. A., Kassam, N., Ruffing, N., Rottman, J. B., Sullivan, N., Choe, H., Sodroski, J., Newman, W., Koup, R. A. & Mackay, C. (1997) J. Exp. Med. 185, 1681–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reynes, J., Portales, P., Segondy, M., Baillat, V., Andre, P., Reant, B., Avinens, O., Couderc, G., Benkirane, M., Clot, J., et al. (2000) J. Infect. Dis. 181, 927–932. [DOI] [PubMed] [Google Scholar]
- 32.Lee, B., Sharron, M., Montaner, L., Weissman, D. & Doms, R. (1999) Proc. Natl. Acad. Sci. USA 96, 5215–5220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.O'Brien, S. J. & Moore, J. P. (2000) Immunol. Rev. 177, 99–111. [DOI] [PubMed] [Google Scholar]
- 34.Clerici, M., Butto, S., Lukwiya, M., Sarasella, M., Declich, S., Trabattoni, D., Pastori, S., Fracasso, C., Fabiani, M., Ferrante, P., et al. (2000) AIDS 14, 2083–2092. [DOI] [PubMed] [Google Scholar]
- 35.Blanpain, C., Lee, B., Tackoen, M., Puffer, B., Boom, A., Libert, F., Sharron, M., Wittamer, V., Vassart, G., Doms, R. W. & Parmentier, M. (2000) Blood 96, 1638–1645. [PubMed] [Google Scholar]
- 36.Farzan, M., Mirzabekov, T., Kolchinsky, P., Wyatt, R., Cayabyab, M., Girard, N. & Girard, C. (1999) Cell 96, 667–676. [DOI] [PubMed] [Google Scholar]
- 37.Paxton, W. A., Neumann, A. U., Kang, S., Deutch, L., Brown, R. C., Koup, R. A. & Wolinsky, S. M. (2001) J. Infect. Dis. 183, 1678–1681. [DOI] [PubMed] [Google Scholar]
- 38.Hammer, S. M. & Pedneault, L. (2000) Antiviral Ther. 5, 23–26. [PubMed] [Google Scholar]
- 39.Baldwin, C. E., Sanders, R. W. & Berkhout, B. (2003) Curr. Med. Chem. 10, 1773–1782. [DOI] [PubMed] [Google Scholar]
- 40.Rimsky, L. T., Shugars, D. C. & Matthews, T. J. (1998) J. Virol. 72, 986–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wei, X., Decker, J. M., Liu, H., Zhang, Z., Arani, R. B., Kilby, J. M., Saag, M. S., Wu, X., Shaw, G. M. & Kappes, J. C. (2002) Antimicrob. Agents Chemother. 46, 1896–1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kazmierski, W., Bifulco, N., Yang, H., Boone, L., DeAnda, F., Watson, C. & Kenakin, T. (2003) Bioorg. Med. Chem. 11, 2663–2676. [DOI] [PubMed] [Google Scholar]
- 43.Aarons, E. J., Beddows, S., Willingham, T., Wu, L. & Koup, R. A. (2001) Virology 287, 382–390. [DOI] [PubMed] [Google Scholar]
- 44.Maeda, Y., Foda, M., Matsushita, S. & Harada, S. (2000) J. Virol. 74, 1787–1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schols, D., Esté, J. A., Cabrera, C. & Clercq, E. D. (1998) J. Virol. 72, 4032–4037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kanbara, K., Soto, S., Tanuma, J. I., Tamamura, H., Gotch, K., Hoshimori, M., Kanamoto, T., Kitano, M., Fujii, N. & Nakashima, H. (2001) AIDS Res. Hum. Retroviruses 17, 615–622. [DOI] [PubMed] [Google Scholar]
- 47.Vreese, K. D., Kofler-Mongold, V., Leutgeb, C., Weber, V., Vermeire, K., Schact, S., Anne, J., Clercq, E. D., Datema, R. & Werner, G. (1996) J. Virol. 70, 689–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gorry, P. R., Taylor, J., Holm, G. H., Mehle, A., Morgan, T., Cayabyab, M., Farzan, M., Wang, H., Bell, J. E., Kunstman, K., et al. (2002) J. Virol. 76, 6277–6292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Trkola, A., Kuhmann, S. E., Strizki, J. M., Maxwell, E., Ketas, T., Morgan, T., Pugach, P., Xu, S., Wojcik, L., Tagat, J., et al. (2002) Proc. Natl. Acad. Sci. USA 99, 395–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mosier, D. E., Picchio, G. R., Gulizia, R. J., Sabbe, R., Poignard, P., Picard, L., Offord, R. E., Thompson, D. A. & Wilken, J. (1999) J. Virol. 73, 3544–3550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Esté, J. A., Cabrera, C., Blanco, J., Gutierrez, A., Bridger, G., Henson, G., Clotet, B., Schols, D. & Clercq, E. D. (1999) J. Virol. 73, 5577–5585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gotoh, H., Yoshimori, M., Kanbara, K., Tamamura, H., Kanamoto, T., Mochizuki, K., Fujii, N. & Nakashima, H. (2001) J. Infect. Chemother. 7, 28–36. [DOI] [PubMed] [Google Scholar]
- 53.Parkin, N., Chappey, C., Maroldo, L., Bates, M., Hellmann, N. S. & Petropoulos, C. J. (2002) J. Acquired Immune Defic. Syndr. 31, 128–136. [DOI] [PubMed] [Google Scholar]
- 54.Falloon, J., Ait-Khaled, M., Thomas, D. A., Brosgart, C. L., Eron, J. J., Jr., Feinberg, J., Flanigan, T. P., Hammer, S. M., Kraus, P. W., Murphy, R., et al. (2002) AIDS 16, 387–396. [DOI] [PubMed] [Google Scholar]
- 55.Tremblay, C. L., Giguel, F., Kollman, C., Guan, Y., Chou, T. C., Baroudy, B. M. & Hirsch, M. S. (2002) Antimicrob. Agents Chemother. 46, 1336–1339. [DOI] [PMC free article] [PubMed] [Google Scholar]