Abstract
The insertion and deletion of U residues at specific sites in mRNAs in trypanosome mitochondria is thought to involve 3′ terminal uridylyl transferase (TUTase) activity. TUTase activity is also required to create the nonencoded 3′ oligo[U] tails of the transacting guide RNAs (gRNAs). We have described two TUTases, RET1 (RNA editing TUTase 1) and RET2 (RNA editing TUTase 2) as components of different editing complexes. Tandem affinity purification-tagged Trypanosoma brucei RET2 (TbRET2) was expressed and localized to the cytosol in Leishmania tarentolae cells by removing the mitochondrial signal sequence. Double-affinity isolation yielded tagged TbRET2, together with a few additional proteins. This material exhibits a U-specific transferase activity in which a single U is added to the 3′ end of a single-stranded RNA, thereby confirming that RET2 is a 3′ TUTase. We also found that RNA interference of RET2 expression in T. brucei inhibits in vitro U-insertion editing and has no effect on the length of the 3′ oligo[U] tails of the gRNAs, whereas down-regulation of RET1 has a minor effect on in vitro U-insertion editing, but produces a decrease in the average length of the oligo[U] tails. This finding suggests that RET2 is responsible for U-insertions at editing sites and RET1 is involved in gRNA 3′ end maturation, which is essential for creating functional gRNAs. From these results we have functionally relabeled the previously described TUT-II complex containing RET1 as the guide RNA processing complex.
U-insertion/deletion RNA editing in trypanosome mitochondria involves the annealing of a guide RNA (gRNA) to an mRNA, endonuclease cleavage at a non-base-paired editing site adjacent to the RNA duplex, addition or deletion of Us to or from the 3′ end of the 5′ cleavage fragment, and religation of the two mRNA fragments (1–4). Each gRNA mediates the editing of 1–5 sites in a 3′ to 5′ polarity, and multiple overlapping gRNAs mediate the editing of an entire domain, also in a 3′ to 5′ polarity (5).
We previously isolated the RNA editing terminal uridylyl transferase (TUTase) 1 (RET1) and showed it to be present both as a free tetramer and as a component of a high molecular weight complex (TUT II) (6). This complex migrates on a native gel at ≈700 kDa and interacts in an RNase-sensitive manner with the larger ligase-containing complex (L-complex) (7). Down-regulation of RET1 expression in Trypanosoma brucei by conditional RNA interference (RNAi) caused a decrease in the steady-state abundance of edited mRNAs without any effect on transcription (6). The core L-complex and associated proteins, capable of both U-insertion and U-deletion editing activity in vitro, was isolated from Leishmania tarentolae mitochondrial extract by tandem affinity purification (TAP) (8), followed by glycerol gradient sedimentation (7). The L-complex contains two adenylatable RNA ligases, REL1 and REL2, three related zinc finger-containing proteins, two proteins with RNase III motifs, two proteins with an AP endonuclease_exonuclease_phosphatase motif, a second 3′ TUTase, which we termed RET2, and two proteins with no identifiable motifs (7). Two additional RNase III-motif proteins and a protein with an S1 domain have been identified in the T. brucei editing complex, whereas two of the L. tarentolae proteins, LC-7c and LC-10, were not observed in the T. brucei complex (9, 10). The L-complex depends solely on protein–protein interactions for stability and exists in a dimeric or tetrameric configuration. The interaction of the L-complex with RET1 and with an RNA-binding protein complex is RNA dependent (11). To dissect the biological roles of the two TUTases in RNA editing, we have compared the effects of RET1 and RET2 down-regulation by RNAi on mitochondrial mRNAs, gRNAs, and on in vitro editing activities of RNA-editing complexes. In this article, we demonstrate that RET2 is indeed a 3′ RNA uridylyltransferase, and is responsible for the insertion of Us into the mRNA, whereas the RET1 enzyme is involved in the formation of the posttranscriptionally added oligo[U] tails of the gRNAs.
Materials and Methods
Cell Cultures, RNAi, and TAP Isolation. Construction of RNAi plasmids and strain manipulation was as described (6). The Leishmania major RET2 with a C-terminal TAP tag was inserted into the pX vector (12) and transfected into L. tarentolae cells. TAP was performed as described, except for the gradient sedimentation step (7).
RNA Analysis. Total RNA was purified by the acid guanidium isothiocyanate method and treated with DNase I. Steady-state levels of mitochondrial mRNA editing and gRNAs were analyzed as described (6). RT-PCR was performed with SuperScript II reverse transcriptase and 3 μg of total RNA, according to the commercial protocol.
Extract Preparation, Glycerol Gradient Sedimentation, and Native Gel Electrophoresis. Purified mitochondria (25 mg of protein per ml) (13) was lysed with 0.3% Triton X-100 in 10 mM Tris·HCl, pH 8.0/125 mM sucrose/10 mM MgCl2/60 mM KCl. The clarified extract (300 μl) was centrifuged on a 10–30% glycerol gradient in the SW41 rotor (Beckman) for 20 h at 30,000 rpm. The fractions were diluted 2-fold with water and electrophoresed on 8–16% native gel (NOVEX, San Diego), or mixed with SDS loading buffer for 10–20% denaturing gradient gel electrophoresis, and the gels were blotted for Western analysis. Sedimentation values were calculated by using catalase (11S), thyroglobulin (19S), and small ribosome subunits from Escherichia coli (30S).
RNA Substrates and in Vitro Precleaved Assay. Gradient fractions obtained from T. brucei mitochondrial extracts were screened for in vitro editing activity and for the presence of the REL1 and REL2 RNA ligases. Fractions 9–10 (of 16 total fractions) were used in the precleaved editing assay. Reactions were performed with 1 μg of total protein in the presence of 0.1 mg/ml acetylated BSA for 45 min with chemically synthesized RNA substrates as described (7). The reactions were preincubated for 15 min before the addition of UTP to 400 μM and ATP to 10 μM. Linear product accumulation for addition of two Us was observed up to 60 min.
Expression and Isolation of T. brucei RET2 (TbRET2). The TbRET2 without the 29 N-terminal amino acids was expressed as a TAP fusion protein in a previously described pX-based vector (7). L. tarentolae cells were transfected ,and clonal drug-resistant lines were selected. The cells were grown to 150 × 106 cells per ml and harvested by centrifugation. Approximately 3 × 1011 cells (4.5 g wet weight) were disrupted in 30 ml of 20 mM Tris·HCl, pH 7.8/400 mM NaCl/2 mM EDTA buffer by a single passage through the French press cell at 1,200 psi (1 psi = 6.89 kPa). Nonidet P-40 detergent was added to 0.3%, and the extract was clarified by centrifugation at 50,000 rpm in the Ti65 rotor (Beckman) for 2 h at 4°C and incubated with 0.4 ml (packed volume) of IgG Sepharose (Amersham Biosciences) for 2 h. All subsequent procedures were as described (8). Recombinant TbRET1 was isolated as in ref. 6. Nucleotide triphosphate (NTP) incorporation assay was performed in a 30-μl reaction containing 20 mM Tris·HCl, pH 7.8/30 mM KCl/10 mM MgCl2/1 mM DTT/0.3 mM of NTP/100 nM of the 5′-labeled 5′ fragment used in the precleaved assay, and 5 nM of enzyme at 27°C for 30 min. The enzyme concentration was determined by comparison of Sypro ruby-stained bands in an SDS gel to bands of a known concentration of BSA, assuming one active center per polypeptide.
Results
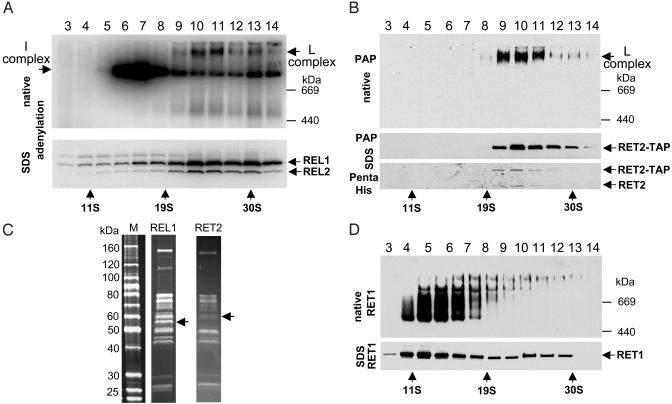
TAP Isolation of Proteins Interacting with RET2. We inserted the L. major RET2 into a TAP expression vector (7) and transfected L. tarentolae cells. The RET2 protein was expressed and targeted to the mitochondrion (data not shown). Mitochondrial extract from a clonal drug-resistant line of transfected cells was fractionated on a glycerol gradient, and each fraction was incubated with [α-32P]ATP, and was further fractionated in native and SDS gels, which were transferred onto nitrocellulose filters. The REL1 and REL2 RNA ligases hydrolyze [α-32P]ATP to covalently bound [32P]AMP and can serve as markers for the RNA ligase-containing L-complex, which sediments ≈20–30 S, and migrates as a discrete high molecular weight band in a native gel (Fig. 1A). We treated the same blots with the peroxidase-antiperoxidase (PAP) reagent, which is specific for the Protein A moiety of the RET2-fusion protein, and retreated the SDS gel blot with a Penta-His antibody (Qiagen, Valencia, CA), which reacts with RET2 because of the fortuitous presence of five adjacent histidine residues in the C-terminal region (7).
Fig. 1.
RET1 and RET2 are components of different complexes. (A) Mitochondrial extract from the RET2-TAP L. tarentolae cell line was fractionated on a glycerol gradient. Each fraction was incubated with [α-32P]ATP and separated on a 4–12% native gel (Upper) or an 8–16% SDS gel (Lower), which were blotted onto filters for PhosphorImager analysis. The L-complex is identified by the labeled REL1 and REL2 ligase bands (Lower) and the single-labeled band (arrow) (Upper). (B) RET2 is present only in the L-complex. The filters were either treated with the PAP reagent to detect the RET2-protein A fusion or with the monoclonal Penta-His antibody (Qiagen) to detect RET2. (C) Comparison of protein compositions of L-complexes purified through TAP-tagged RET2 TUTase or REL1 RNA ligase. Arrows indicated positions of tagged proteins. (D) Distribution of RET1 in gradient. Filter A was treated with polyclonal antibodies against RET1.
The TAP-tagged RET2, as detected by native and denaturing gel analysis of gradient fractions, is found solely in the L-complex (Fig. 1B), which is unlike the situation with the TAP-tagged REL1, which we previously found (7), showed a substantial amount in the 10–12 S region, in addition to the L-complex region.
The presence of both the tagged RET2 and the endogenous RET2 (Penta-His panel of Fig. 1B) in the L-complex and in the RET2-TAP pulldown (see Fig. 1C below) is consistent with a dimeric or tetrameric organization of the L-complex, as was also concluded previously by using the TAP-tagged REL1 protein (7).
Double-affinity purification of the tagged RET2 and interacting proteins yielded a set of proteins very similar to those previously obtained by using the TAP-tagged REL1 protein (ref. 7 and Fig. 1C), further substantiating the conclusion that RET2 is an integral component of the L-complex, which is involved in both U-insertion and U-deletion editing.
Fig. 1D shows that only a minor fraction of RET1 comigrates with the TAP-tagged L-complex. This result is consistent with our previous finding of an unstable RNA-dependent interaction of RET1 with REL1 and REL2 RNA ligases (7).
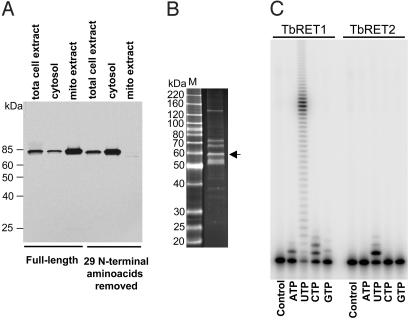
RET2 Is a 3′ Uridylyl Transferase That Adds a Single U Residue. The high degree of sequence similarity between RET1 and RET2 (7) in concert with the “loss-of-function” RNAi results presented below, strongly suggest that RET2 is a uridylyl transferase, but multiple attempts to express soluble and active RET2 protein in a variety of bacterial and yeast systems were unsuccessful. However, when expressed in L. tarentolae, both the L. major RET2 (Fig. 1B) and TbRET2 (data not shown) preproteins were targeted to mitochondria and incorporated into the L-complex. This finding prompted us to investigate whether L. tarentolae cells themselves can be used as a source of soluble and active editing proteins by cytosolic expression of polypeptides with the mitochondrial importation peptide sequences removed. We have no direct information on the signal peptide cleavage site for TbRET2, but the mitoprot and signalp programs both indicate a cleavage site at 25 amino acids. To be certain the signal sequence was deleted, we removed 29 amino acids from the N terminus by recombinant PCR, and we also inserted an ATG codon for initiation of translation. L. tarentolae cells expressing TAP-tagged TbRET2 proteins with and without the putative mitochondrial signal peptide were disrupted under hypotonic conditions (“total-cell extract”), and were separated by centrifugation into a soluble fraction (“cytosol”) and a crude mitochondria fraction. Mitochondria were purified from the latter by flotation in a renografin density gradient, and mitochondrial proteins were extracted as described for the TAP purification. The fractionation into cytosol and mitochondria produced little cross contamination, as shown by analysis of the mitochondrial matrix marker enzyme, glutamate dehydrogenase, and the cytosolic marker protein, tubulin (data not shown).
Western analysis of the expressed TbRET2 protein with the PAP-reagent (Fig. 2A) showed that the removal of the mitochondrial signal had no effect on the expression level or stability of the protein. In addition, the removal of the mitochondrial signal sequence, as predicted, prevented importation into the mitochondrion, and the TbRET2-TAP protein was soluble. The tagged protein was purified by two consecutive IgG Sepharose and calmodulin agarose affinity columns (Fig. 2B). Unexpectedly, five additional proteins copurified with tagged TbRET2 from the total Leishmania cell extract, notwithstanding the rigorous TAP procedure. A control experiment in which cells were transfected with a plasmid containing only the TAP cassette yielded no protein bands or TUTase activity (data not shown). These proteins represent a much simpler set than L-complex (Fig. 1C) and may reflect high-affinity interactions with mitochondrial proteins released during cell breakage or with mitochondrial preproteins in the cytosol, but this remains to be determined. Nevertheless, the RET2 protein represents a major stained band in the isolated material.
Fig. 2.
3′ RNA uridylyl transferase activity of TbRET2. (A) Deletion of mitochondrial signal peptide results in the cytosolic localization of the TbRET2. Ten micrograms of protein from total cell extract, soluble fraction, and the S100 mitochondrial extract were fractionated on an 8–16% SDS gel, transferred onto nitrocellulose membrane, and treated with the PAP reagent to detect protein A fusion proteins. (B) Isolation of recombinant TbRET2 from L. tarentolae. The fraction eluted from the calmodulin column (see Materials and Methods) was analyzed on a 10–20% SDS gel followed by staining with Sypro ruby. Arrow indicates position of the tagged TbRET2. (C) Uridylyl transferase activity of recombinant TbRET1 and TbRET2 enzymes. The 5′ end-labeled 5′ fragment used in the precleaved assay was incubated with the respective enzyme in the presence of 0.3 mM ribonucleotide triphosphates, and the products were separated on a 14% polyacrylamide/urea sequencing gel. Control, no NTP added.
In an in vitro assay for 3′ transferase activity, the purified TbRET2-TAP material incorporates mostly one U residue with traces of the +2 and +3 products and is specific for UTP (Fig. 2C). Under identical conditions, the recombinant RET1 protein produces a ladder of products (Fig. 2C).
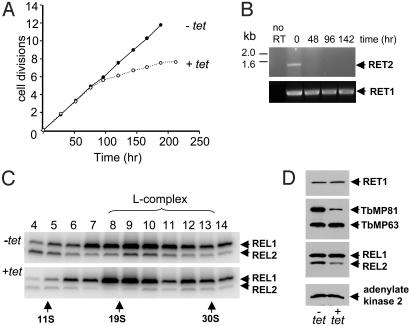
RNAi Down-Regulation of RET2 Expression. To analyze the specific functions of the two TUTases, we decided to perform conditional RNAi down-regulation of RET2 expression, and to compare the effect on the length distribution of the 3′ oligo[U] tail of gRNAs, and on the in vitro editing activities to that of cells in which RET1 expression was down-regulated. Because Leishmania cells do not exhibit RNAi, it was necessary to use T. brucei to perform conditional down regulation of RET2 expression. However, the results should be applicable to L. tarentolae because the T. brucei and L. tarentolae RET2 TUTases show 67% amino acid identity and the TbRET2 is also a component of a high molecular weight editing complex in these cells (10). T. brucei strain 29–13 procyclic cells (14), expressing T7 RNA polymerase and tetracycline repressor, were transfected with a plasmid containing two fragments of RET2 in a head-to-head configuration under control of a single tetracycline-regulatable T7 promoter (15). Induction of RNAi of RET2 expression with tetracycline produced growth inhibition by ≈100 h (Fig. 3A). Degradation of RET2 mRNA within 2 days of RNAi induction was observed by RT-PCR analysis, whereas the RET1 mRNA was not affected (Fig. 3B).
Fig. 3.
Effects of RNAi down-regulation of TbRET2 expression in procyclic T. brucei on cell growth and stability of the L-complex. (A) RNAi was induced by addition of 1 μg/ml tetracycline. Cell growth is affected after ≈100 h of RNAi. (B) RT-PCR analysis of RET1 and RET2 mRNAs during RET2 RNAi. (C) Glycerol gradients of mitochondrial extract from uninduced and 3-day RNAi-induced cells. Fractions were labeled with [α-32P]ATP and separated in an SDS gel. The location of the L-complex is indicated. (D) Mitochondrial extracts from uninduced or 3-day RNAi-induced cells were fractionated in an SDS gel, and the blot reacted with antisera against the indicated proteins.
Stability of L-complex Is Unaffected by Down-Regulation of RET2. Interestingly, despite the down-regulation of RET2 expression, there was no major effect on the overall stability of the L-complex, as shown by sedimentation of mitochondrial extract from cells induced for RET2 RNAi for 3 days (Fig. 3C). In this experiment we used adenylation of REL1 and REL2 to localize the L-complex in the gradient, and it appears that there is also a correlated loss of adenylatable REL2 in the L-complex fractions (Fig. 3C Lower). Western analysis of total mitochondrial extract confirmed the decrease in relative abundance of REL2, and also TbMP81, and the lack of effect on REL1, TbMP63, or adenylate kinase, an unrelated mitochondrial protein (Fig. 3D). This correlation suggests a regulated interaction of RET2, REL2, and TbMP81 (the homologue of TbMP81 in L. major is LC-1). These proteins are components of the REL2-subcomplex within the core L-complex (16). We showed previously (7) that overexpression of REL1 in L. tarentolae produces a subcomplex with LC-3 (TbMP99 homologue), LC-4 (TbMP63 homologue), and LC-5 (TbMP46 homologue) (10). In addition, we have recently reported that REL1 and TbMP63 are also coregulated (17), as was also found for RNAi down-regulation of TbMP63 (18). These data suggest that the REL1 and REL2 subcomplexes are structurally separated within the L-complex as tightly bound, defined groups of proteins. It is also possible that the formation of subcomplexes precedes the assembly of the L-complex, which would explain the formation of an unincorporated REL1-subcomplex as a result of REL1-TAP overexpression in L. tarentolae (7).
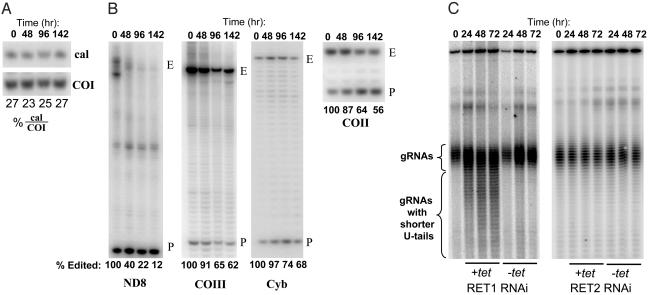
Inhibition of RNA Editing in Vivo by Down-Regulation of RET2. We had shown that down-regulation of the RET1 by RNAi caused a selective decrease in the steady-state abundance of edited mRNA transcripts, indicating an inhibition of RNA editing (6). A similar effect on the abundance of edited mRNA transcripts was observed on RNAi down-regulation of RET2 (Fig. 4B). A control primer extension experiment (Fig. 4A) showed the lack of effect on the cytosolic calmodulin mRNA, or on the never-edited COI mRNA. The magnitude of the inhibition of editing was transcript specific, and reached a maximum of ≈90% in the case of the ND8 mRNA. In general, the extent of inhibition of in vivo editing was less than that of RET1 RNAi, except for the COII mRNA, which contains the gRNA at the 3′ end in cis. The inhibition of editing by RET1 and RET2 RNAi could be either a direct effect on the addition of Us to the 3′ end of the 5′ mRNA cleavage fragment, or an indirect effect because of interference with 3′ end processing of gRNAs.
Fig. 4.
Effect of RET1 and RET2 RNAi on in vivo mRNA editing and on length of gRNA oligo[U] tails. (A) The never-edited COI and cytosolic calmodulin mRNAs were analyzed in the same reactions as internal loading controls. (B) The relative abundance of fully edited and preedited ND8, COIII, Cyb, and COII transcripts was analyzed by primer extension. E, fully edited; P, preedited. Relative percentages of edited transcripts are shown below. (C) gRNAs were 5′ labeled with [α-32P]GTP in the presence of guanylyltransferase and separated on 12% polyacrylamide/urea gel. (Left) RET1 RNAi. (Right) RET2 RNAi. The band at the top of the gel is a cytosolic RNA that labels with [α-32P]GTP and is used as a loading control.
Down-Regulation of RET1 Affects Length Distribution of gRNA 3′ Oligo[U] Tails, Whereas Down-Regulation of RET2 Has No Effect. Total RNA from cells after 72 h of RET1 or RET2 RNAi induction was incubated with [α-32P]GTP and guanylyltransferase (1, 5) and fractionated on a sequencing gel. This procedure specifically labels RNAs with 5′ tri- or diphosphates, such as gRNAs. An unidentified cytoplasmic RNA at the top of the gel is also labeled and was used as a loading control (Fig. 4C). Under denaturing conditions, the total gRNA population migrates as a tight set of bands in the 65–75 nucleotide range. The appearance of a population of smaller gRNAs due to a decrease in the length of the oligo[U] tails was detectable after 1 day of RNAi of RET1 (Fig. 4C Left), whereas there was no change in the length distribution of oligo[U] tails after RNAi of RET2 (Fig. 4C Right). PhosphorImager analysis of the lanes corresponding to 72 h of RET1 RNAi induction shows that in the control tetracycline (–tet) lane ≈90% of the radioactivity is located in the region indicated as gRNAs (Fig. 4C), and only 10% corresponds to the region with molecules shortened by 1–20 bases, which covers the average length of the oligo[U] tail (19). Down-regulation of RET1 expression shifted this balance to ≈60% of gRNAs having shortened oligo[U] tails. These data indicate that the RET1 TUTase is involved with the addition of Us to the 3′ ends of the gRNAs and also shows that a mature 3′ oligo[U] tail is essential for RNA editing.
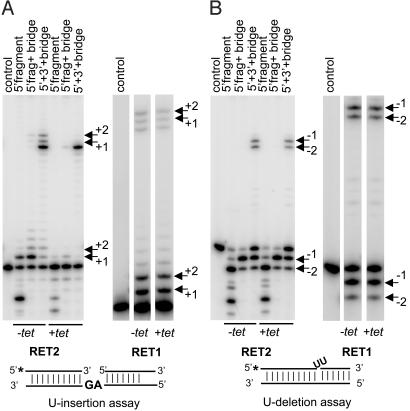
Differential Effects of RET1 and RET2 Down-Regulation on in Vitro Precleaved U-Insertion Editing. We also analyzed the effects of RET1 and RET2 RNAi on in vitro editing. The L-complex was partially purified by glycerol gradient sedimentation of mitochondrial extract from cells after 3 days of RNAi induction and used to measure in vitro editing activity in precleaved assays (Fig. 5). This assay uses an RNA substrate consisting of the 5′ and 3′ mRNA cleavage fragments annealed to a bridge RNA, which either has two guiding nucleotides for U-insertions or no guiding nucleotides for two U-deletions (20). In the case of RET1 RNAi, there was an ≈15% decrease in the production of +2U edited RNA (Fig. 5A) and no detectable effect on the production of –2U edited RNA (Fig. 5B). In the case of RET2 RNAi, there was a complete inhibition of U-insertion activity (Fig. 5A), but no detectable effect on U-deletion activity (Fig. 5B). This dramatic difference between the effects of down-regulation of RET1 and RET2 on U-insertion editing suggests that RET2 is responsible for the insertion of Us into mitochondrial mRNA in U-insertion editing. Combined with the above results, we conclude that the RET1 effect on editing is indirect and probably involves the gRNA-oligo[U] tail.
Fig. 5.
Effect of RET1 RNAi and RET2 RNAi on in vitro precleaved editing activity. (A) RNAi was induced for 3 days, and mitochondrial extract was fractionated in a glycerol gradient. The L-complex fractions were used for the U-insertion assay. The RNA substrates are diagrammed. Positions of extended 5′ fragments and the corresponding ligation products are indicated by arrows. (B) U-deletion assay. See A for description. The 5′ fragments with one or two Us removed and corresponding ligation products are indicated by arrows.
Discussion
The RET2 protein, which is a component of the REL2-subcomplex, has the characteristic nucleotidyl transferase and poly(A) polymerase motifs present in the RET1 TUTase (7). To confirm this enzymatic identification, TAP-tagged TbRET2 protein with the mitochondrial signal peptide removed was overexpressed in L. tarentolae and the RET2 protein, together with several interacting proteins purified by double-affinity chromatography. The recombinant TbRET2 exhibits a U-specific 3′ TUTase activity, which differs from that of the RET1 TUTase by the addition of a single U residue at the same UTP concentration at which RET1 adds a ladder of U residues.
Down-regulation of expression of TbRET2 by RNAi causes a complete inhibition of in vitro U-insertion editing without any effect on U-deletion editing, and has no effect on the 3′ oligo[U] tails of the gRNAs. The extent of inhibition of editing in vivo varies from mRNA to mRNA, probably as a consequence of differential RNA stability. Down-regulation of RET1 expression, on the other hand, affects the length of the oligo[U] tails and has much less effect on in vitro U-insertion editing. We conclude that RET2 is responsible for the addition of Us to the 3′ end of the 5′ mRNA cleavage fragments in U-insertion editing, and that the RET1 TUTase is involved in the formation of the gRNA 3′ oligo[U] tails. We also conclude that there is a functional requirement for the 3′ oligo[U] tail of the gRNAs, although the exact role remains to be established. In view of these results, we propose to label the ≈700 kDa RET1-containing TUT-II complex the guide RNA processing (GP)-complex (6).
The GP-complex interacts with the L-complex through RNA (7), and we have previously described a third complex with RNA annealing activity that also interacts with the L-complex and the GP-complex through RNA (11). The nature of the RNA linker is unknown, but bound gRNA is a likely candidate. This model is almost certainly incomplete, because there are a number of proteins, such as the putative RNA helicase, mHEL61 (21), the mRNA-binding protein, REAP1 (22), and the oligo[U]-binding proteins RBP16 (23), and TBRGG1 (24), which are also possibly involved in editing.
Acknowledgments
We thank K. Stuart for the monoclonal antibodies against TbMP63 and TbMP81; G. Gao for antibodies against TbREL1 and TbREL2; and D. Nierlich for antibodies against L. tarentolae adenylate kinase. The TAP plasmid was obtained from CellZome (Heidelberg). This work was supported in part by a research grant from the National Institutes of Health (L.S.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: TUTase, terminal uridylyl transferase; RET, RNA editing TUTase; TbRET, Trypanosoma brucei RET; gRNA, guide RNA; L-complex, ligase-containing complex; TAP, tandem affinity purification; RNAi, RNA interference.
See commentary on page 10583.
References
- 1.Blum, B., Bakalara, N. & Simpson, L. (1990) Cell 60, 189–198. [DOI] [PubMed] [Google Scholar]
- 2.Seiwert, S. D. & Stuart, K. (1994) Science 266, 114–117. [DOI] [PubMed] [Google Scholar]
- 3.Seiwert, S. D., Heidmann, S. & Stuart, K. (1996) Cell 84, 831–841. [DOI] [PubMed] [Google Scholar]
- 4.Cruz-Reyes, J. & Sollner-Webb, B. (1996) Proc. Natl. Acad. Sci. USA 93, 8901–8906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maslov, D. A. & Simpson, L. (1992) Cell 70, 459–467. [DOI] [PubMed] [Google Scholar]
- 6.Aphasizhev, R., Sbicego, S., Peris, M., Jang, S. H., Aphasizheva, I., Simpson, A. M., Rivlin, A. & Simpson, L. (2002) Cell 108, 637–648. [DOI] [PubMed] [Google Scholar]
- 7.Aphasizhev, R., Aphasizheva, I., Nelson, R. E., Gao, G., Simpson, A. M., Kang, X., Falick, A. M., Sbicego, S. & Simpson, L. (2003) EMBO J. 22, 913–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Puig, O., Caspary, F., Rigaut, G., Rutz, B., Bouveret, E., Bragado-Nilsson, E., Wilm, M. & Seraphin, B. (2001) Methods 24, 218–229. [DOI] [PubMed] [Google Scholar]
- 9.Stuart, K., Panigrahi, A. K., Schnaufer, A., Drozdz, M., Clayton, C. & Salavati, R. (2002) Philos. Trans. R. Soc. London B 357, 71–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Panigrahi, A. K., Schnaufer, A., Ernst, N. L., Wang, B., Carmean, N., Salavati, R. & Stuart, K. (2003) RNA 9, 484–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aphasizhev, R., Aphasizheva, I., Nelson, R. E. & Simpson, L. (2003) RNA 9, 62–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.LeBowitz, J. H., Coburn, C. M., McMahon-Pratt, D. & Beverley, S. M. (1990) Proc. Natl. Acad. Sci. USA 87, 9736–9740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Braly, P., Simpson, L. & Kretzer, F. (1974) J. Protozool. 21, 782–790. [DOI] [PubMed] [Google Scholar]
- 14.Wirtz, E., Leal, S., Ochatt, C. & Cross, G. A. (1999) Mol. Biochem. Parasitol. 99, 89–101. [DOI] [PubMed] [Google Scholar]
- 15.LaCount, D. J., Bruse, S., Hill, K. L. & Donelson, J. E. (2000) Mol. Biochem. Parasitol. 111, 67–76. [DOI] [PubMed] [Google Scholar]
- 16.Drozdz, M., Palazzo, S. S., Salavati, R., O'Rear, J., Clayton, C. & Stuart, K. (2002) EMBO J. 21, 1791–1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gao, G. & Simpson, L. (2003) J. Biol. Chem. 278, 27570–27574. [DOI] [PubMed] [Google Scholar]
- 18.Huang, C. E., O'Hearn, S. F. & Sollner-Webb, B. (2002) Mol. Cell. Biol. 22, 3194–3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blum, B. & Simpson, L. (1990) Cell 62, 391–397. [DOI] [PubMed] [Google Scholar]
- 20.Igo, R. P., Palazzo, S. S., Burgess, M. L., Panigrahi, A. K. & Stuart, K. (2000) Mol. Cell. Biol. 20, 8447–8457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Missel, A., Souza, A. E., Nörskau, G. & Göringer, H. U. (1997) Mol. Cell. Biol. 17, 4895–4903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Madison-Antenucci, S., Sabatini, R. S., Pollard, V. W. & Hajduk, S. L. (1998) EMBO J. 17, 6368–6376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hayman, M. L. & Read, L. K. (1999) J. Biol. Chem. 274, 12067–12074. [DOI] [PubMed] [Google Scholar]
- 24.Vanhamme, L., Perez-Morga, D., Marchal, C., Speijer, D., Lambert, L., Geuskens, M., Alexandre, S., Ismaïli, N., Göringer, U., Benne, R., et al. (1998) J. Biol. Chem. 273, 21825–21833. [DOI] [PubMed] [Google Scholar]