Abstract
ATRX syndrome is characterized by X-linked mental retardation associated with α-thalassemia. The gene mutated in this disease, ATRX, encodes a plant homeodomain-like finger and a SWI2/SNF2-like ATPase motif, both of which are often found in chromatin-remodeling enzymes, but ATRX has not been characterized biochemically. By immunoprecipitation from HeLa extract, we found that ATRX is in a complex with transcription cofactor Daxx. The following evidence supports that ATRX and Daxx are components of an ATP-dependent chromatin-remodeling complex: (i) Daxx and ATRX can be coimmunoisolated by antibodies specific for each protein; (ii) a proportion of Daxx cofractionates with ATRX as a complex of 1 MDa by gel-filtration analysis; (iii) in extract from cells of a patient with ATRX syndrome, the level of the Daxx–ATRX complex is correspondingly reduced; (iv) a proportion of ATRX and Daxx colocalize in promyelocytic leukemia nuclear bodies, with which Daxx had previously been located; and (v) the ATRX complex displays ATP-dependent activities that resemble those of other chromatin-remodeling complexes, including triple-helix DNA displacement and alteration of mononucleosome disruption patterns. But unlike the previously described SWI/SNF or NURD complexes, the ATRX complex does not randomize DNA phasing of the mononucleosomes, suggesting that it may remodel chromatin differently. Taken together, the results suggest that ATRX functions in conjunction with Daxx in a novel chromatin-remodeling complex. The defects in ATRX syndrome may result from inappropriate expression of genes controlled by this complex.
Keywords: SWI/SNF
Chromatin-remodeling complexes play major roles in regulation of gene expression in eukaryotes (1, 2). These complexes can modify chromatin structure through two distinct mechanisms. One is through covalent modification, including methylation, phosphorylation, and acetylation. The other is through noncovalent mechanisms, which include ATP-dependent chromatin remodeling. We and others have previously purified and characterized several ATP-dependent chromatin-remodeling complexes of the human SWI/SNF family (3–6). In attempts to characterize additional remodeling complexes, we reasoned that because all known ATP-dependent remodeling complexes contain a subunit with a SWI2/SNF2-like ATPase motif, human proteins containing this motif might reveal unique remodeling complexes. A previous study based on this strategy indeed identified the NURD complex (7). Here, we characterize a complex containing ATRX, a protein initially identified by mutations in patients with ATRX syndrome (8).
Mutations in the ATRX gene are now known to cause several X-linked mental retardation syndromes. The phenotypes include facial dysmorphism, urogenital defects, and α-thalassaemia (resulting from reduced α-globin expression) (9). However, the ATRX protein has not been biochemically characterized. As a result, the etiology of ATRX syndrome is poorly understood. Several lines of evidence hinted that ATRX protein may be part of a chromatin-remodeling complex. First, ATRX protein not only has a SWI/SNF2-type ATPase/helicase motif but also has a plant homeodomain-like zinc finger (10), both of which have been found in molecules that modify chromatin structure. Second, ATRX in nuclear extracts fractionates as a complex between 700 and 2,000 kDa (11), a size similar to that of other SWI/SNF complexes. Third, ATRX localizes at pericentromeric heterochromatin (12) and has been identified in yeast two-hybrid screens by its interaction with the heterochromatin protein HP1 as well as a polycomb group protein EZH2 (12–14). Fourth, ATRX mutations have been correlated with changes in DNA methylation patterns at several genomic loci (15). Here, we demonstrate that ATRX forms a complex with a transcription cofactor, Daxx, and this complex displays chromatin-remodeling activities. The results provide a step toward understanding the physiological function of ATRX.
Materials and Methods
Cell Culture. HeLa cells were purchased from the National Cell Culture Center (Minneapolis). The cell line derived from an ATRX patient carries an R37X mutation (16).
Antibodies (Abs). An affinity-purified polyclonal Ab, FXNP5 (NP5), and an ATRX mAb (23c) have been described (17). Two ATRX Abs, D19 and C16, were obtained from Santa Cruz Biotechnology. Two additional rabbit Abs were raised against fusion proteins also containing maltose-binding protein (New England Biolabs) and regions of ATRX (amino acids 365–473 and 2239–2492). Two different Daxx Abs, SC7152 and SC7001, were obtained from Santa Cruz Biotechnology.
Nuclear Extract Fractionation and Immunoprecipitation. HeLa nuclear extract was prepared as described (4). ATRX-associated complex was isolated from HeLa nuclear extract by using an immunoprecipitation protocol (C. S. Lee, Y.X., X. Zhang, and W.W., unpublished data). This protocol and methods for mass spectrometry (MS) analysis, have been described in another study (18). The Superose 6 gel-filtration analysis has been described (19). After fractionation, the ATRX peak fractions were collected and incubated with or without ethidium bromide (100 μg/ml) for 1 h before immunoprecipitation.
Enzymatic Assays. The mononucleosome disruption assay (20, 21) and ATPase assay (22) followed the same procedure used for human SWI/SNF complexes. One modification of the ATPase assay is that the Pi is separated from [γ-32P]ATP by TLC, and the radioactivity was quantitated by using a PhosphorImager. The triple-helix displacement assay used the same protocol and substrates used for RSC and STH1 (23). The double-helix displacement assay used assay conditions and substrates identical to those described (24).
Immunofluorescence. Immunofluorescence experiments followed a previous procedure (12), except that cells were fixed in 100% methanol and then air-dried before immunostaining. Primary Abs included: mouse monoclonal anti-ATRX (23c), rabbit polyclonal anti-Daxx (Ab-1, Oncogene Research Products), rabbit polyclonal anti-promyelocytic leukemia (PML; kindly provided by Paul Freemont, Imperial College, London). All cells were counterstained with 4′,6-diamidino-2-phenylindole (Sigma).
Results
Identification of an ATRX-Associated Complex. Our initial attempts to purify the ATRX-associated complexes used a combination of conventional and immunoaffinity chromatography, a strategy that successfully isolated the human SWI/SNF and NURD complexes. However, such approaches obtained ATRX as a single polypeptide with no associated proteins [data not shown; Yi Zhang's group has independently obtained the same result (Y. Zhang, personal communication)]. The ATRX-associated complex was later found to be unstable in salt solutions of 0.5 M or higher (see Fig. 4e). Fractionation of nuclear extract often resulted in partial or complete dissociation of the ATRX complex. For this reason, we adopted an optimized immunoprecipitation method (C. S. Lee, Y.X., X. Zhang, and W.W., unpublished data) to directly isolate ATRX-associated complexes from HeLa nuclear extract. This method has been used successfully to isolate complexes involved in Bloom syndrome and Fanconi anemia (18).
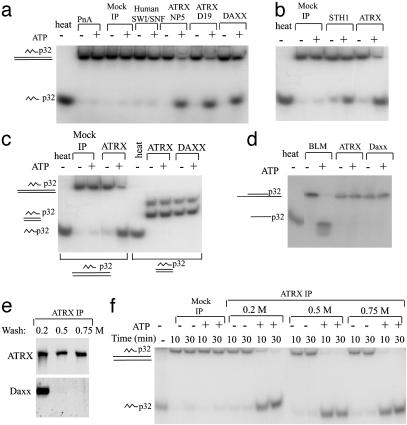
Fig. 4.
The ATRX complex has an ATP-dependent triple-helix displacement activity. (a and b) Autoradiographs showing that the ATRX–Daxx complex displaces a triple helix in the presence of ATP. The positions of the labeled triple-helix substrate and the displaced third strand in the gel are depicted at the left. The complexes isolated by different Abs to ATRX, Daxx, and human SWI/SNF are shown at the top. Polypeptides isolated by mock immunoprecipitation by using either protein A beads (PnA) or preimmune serum (mock IP) are also indicated. Human SWI/SNF in our hands does not show significant activity in this assay. To serve as a positive control, recombinant STH1 (a generous gift of B. Cairns, Huntsman Cancer Institute, University of Utah School of Medicine, Salt Lake City) was used. (c) Autoradiograph showing that the ATRX–Daxx complex does not displace a blunt triple helix. The substrates used are indicated at the bottom. The substrates used in the blunt triplex assay contain two bands. The major band (with the faster mobility) represents the blunt triplex. The minor band (with slower mobility) represents a triplex substrate with one blunt end and one overhang end of 17-bp double-stranded DNA. This overhang was designed to help annealing of the double-stranded DNA and was supposed to be removed by restriction digestion, but it was not completely removed in the current experiment. (d) Autoradiograph showing the results of a double-helix displacement assay. The duplex substrate and the displaced oligo are illustrated at the left. The BLM (Bloom Syndrome gene product) DNA helicase complex was used as a positive control (18). (e) Immunoblot showing that Daxx was completely removed by washing with buffer containing 0.5 or 0.75 M salt, as shown at the top. (f) Autoradiograph showing that dissociation of Daxx does not affect triplex unwinding activity of ATRX.
Six major polypeptides were immunoisolated by a polyclonal Ab against ATRX (Fig. 1a, lane 3). The 300-kDa polypeptide was identified as ATRX by both MS analysis and immunoblotting (Fig. 1b). A minor polypeptide of 180 kDa was also identified as ATRX, which likely represents an alternatively spliced product (R.G. and D.H., unpublished data). The 110-kDa polypeptide was identified as Daxx (Fig. 1 a and b), a protein previously shown to interact with multiple transcription factors (25).
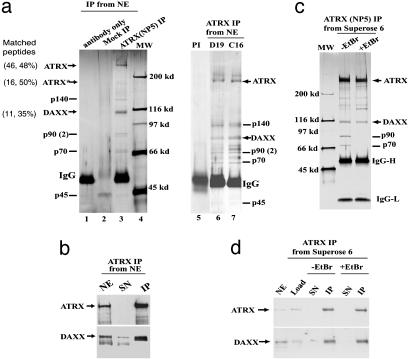
Fig. 1.
Purification of an ATRX-associated complex from HeLa nuclear extract. (a) Silver-stained SDS gels showing polypeptides immunoisolated by three different ATRX Abs (NP5, D19, and C16) from nuclear extract. Mock immunoprecipitation was done by using either protein A beads alone (lane 2) or a preimmune (PI) serum (lane 5). MS has been used for identification of ATRX and its associated polypeptides in all three preparations. The number of peptides that matched the indicated protein and the percentage of these peptides among total peptides obtained from matrix-assisted laser desorption ionization–time-of-flight analysis are shown in parentheses for ATRX and Daxx. The other polypeptides (marked by lines) appeared to either loosely associate with ATRX or associate with ATRX through DNA by subsequent analysis. (b) Immunoblot confirming the presence of Daxx in the polypeptides immunoisolated by ATRX Ab. NE, nuclear extract; SN, supernatant; IP, immunoprecipitate. The lower Daxx band in nuclear extract could represent a posttranslationally modified version of Daxx that associates poorly with SDS. (c) Silver-stained gel showing polypeptides immunoisolated by an ATRX Ab from the ATRX peak fractions after Superose 6 fractionation of nuclear extract (Fig. 2c). The presence or absence of ethidium bromide (EtBr) is indicated. Note that p90 and p70 polypeptides are lost in IP in the presence of EtBr, hinting that they may associate with ATRX through DNA. Also, Daxx becomes substoichiometric compared to ATRX, which could be caused by partial dissociation of the ATRX–Daxx complex during Superose 6 fractionation. (d) Immunoblot showing that association between ATRX and Daxx is not through DNA.
Daxx and other copurifying polypeptides were further analyzed to determine their possible association with ATRX. All of them can be isolated by four additional ATRX Abs against different regions of ATRX (Fig. 1a, lanes 6 and 7, and data not shown) but not by protein A beads alone (Fig. 1a, lane 2) or a preimmune serum (Fig. 1a, lane 5), suggesting that they are not isolated because of Ab crossreactivity. Immunoprecipitation by using ATRX peak fractions from Superose 6 fractionation of nuclear extract (see Fig. 2c) obtained fewer major polypeptides, including ATRX, Daxx, p90, and p70 (Fig. 1c). To rule out the possibility that these polypeptides may associate with ATRX through DNA, immunoprecipitation was performed in the presence of ethidium bromide, a DNA-intercalating drug that can dissociate proteins from DNA and has often been used to identify DNA-independent protein associations (26). The immunoprecipitation yielded Daxx but not p90 and p70 (Fig. 1 c and d), suggesting that Daxx, but not the latter two proteins, forms a DNA-independent complex with ATRX.
Fig. 2.
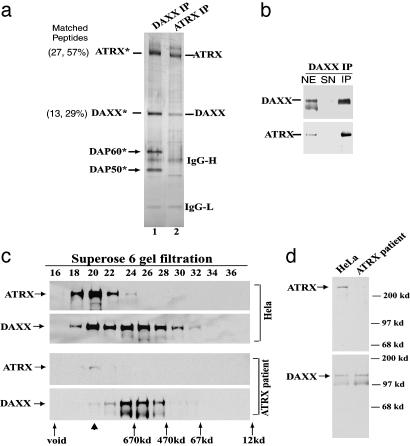
Daxx and ATRX form a complex, and the level of this complex is significantly decreased in a ATRX patient cell line. (a) Silver-stained SDS gel showing the polypeptides immunoisolated by a Daxx Ab compared to those by the ATRX Ab. All polypeptides marked by an asterisk have been identified by MS. The number of peptides that match the indicated protein and the percentage of these peptides among total peptides obtained from matrix-assisted laser desorption ionization–time-of-flight analysis are shown in parentheses for ATRX and Daxx. (b) Immunoblot confirming the presence of ATRX in the polypeptides isolated by the Daxx Ab. (c) Immunoblot showing the Superose 6 gel-filtration profile of ATRX and Daxx in nuclear extracts prepared from either HeLa cells (Upper) or a cell line derived from an ATRX patient (Lower). The peaks corresponding to proteins with known molecular masses are denoted at the bottom. Note that the peak of the ATRX–Daxx complex (fraction 20, indicated by an arrowhead) is significantly reduced in the ATRX patient cell line. (d) Immunoblot analysis showing the levels of ATRX (Upper) and Daxx (Lower) in the ATRX patient cell line in comparison to those in HeLa cells.
ATRX Forms a Complex with Transcriptional Coactivator Daxx. Daxx is present in nearly equimolar amounts with ATRX among polypeptides isolated by ATRX Ab directly from nuclear extract (Fig. 1a). The level of Daxx is also significantly reduced in the supernatant after immunoprecipitation by the ATRX Ab (Fig. 1b). The results suggest that a significant proportion of Daxx in the nuclear extract forms a stable complex with ATRX.
Daxx was originally identified by a yeast two-hybrid screen designed to detect proteins interacting with the Fas receptor, which localizes at the cytoplasmic membrane and mediates apoptosis (27). Immunofluorescence studies later showed that the endogenous Daxx is exclusively a nuclear protein (28), and its function is likely to modulate transcription (29, 30). Consistent with a primary role in transcription, Daxx has been identified in two-hybrid screens for its interaction with multiple transcription factors, DNA methyltransferase I, histone deacetylases, and core histones (30–37). However, endogenous Daxx has not been purified by unbiased biochemical approaches. Consequently, basic questions regarding Daxx remain unanswered, including the number and composition of Daxx complexes that exist in a given cell type.
To assess independently whether Daxx and ATRX form a complex, we used an unbiased approach to isolate Daxx-associated polypeptides. Immunopurification by using a Daxx Ab isolated four major polypeptides in near equimolar ratios (Fig. 2a). Immunopurification was highly efficient because little Daxx remained in the supernatant after the purification (Fig. 2b), suggesting that most of the endogenous Daxx in the extract was isolated by this approach. The 110-kDa polypeptide was identified as Daxx by both MS and immunoblotting analysis (Fig. 2 a and b). Similarly, the 300-kDa polypeptide was identified as ATRX. The findings that ATRX and Daxx can be immunoisolated in near equimolar amounts by Abs specific for each protein strongly suggest that they are components of one complex. In addition, after immunopurification by the Daxx Ab, the amount of ATRX in the supernatant was almost completely depleted, suggesting that most, if not all, ATRX is in a complex with Daxx in HeLa nuclear extracts (Fig. 2b).
ATRX and Daxx Cofractionate as a 1-MDa Complex. ATRX in HeLa nuclear extract fractionated with a single peak in Superose 6 gel-filtration analysis, corresponding to a complex of 1 MDa (Fig. 2c). This result is consistent with a previous report that ATRX fractionates in a complex of 0.7–2 MDa (11). Daxx fractionated in two peaks: one of them overlaps with that of ATRX (at fraction 20) and could correspond to the ATRX–Daxx complex. The other one is ≈700 kDa (fraction 25) and should represent a complex without ATRX. Thus, most ATX in the extract is present in the ATRX–Daxx complex, but a significant proportion of Daxx can form a complex independent of ATRX. The results are in agreement with the immunoprecipitation data above: not all Daxx was immunodepleted by the ATRX Ab (Fig. 1d), but almost all ATRX was immunodepleted by the Daxx Ab (Fig. 2b).
The Level of the ATRX–Daxx Complex Is Significantly Decreased in an ATRX Patient Cell Line. If two proteins are components of a complex, one would expect that when one protein is absent, the complex containing the other protein should become smaller when it is fractionated by gel-filtration chromatography. This experimental strategy has previously been used to demonstrate that five SWI/SNF proteins are parts of one complex (38). Here, we used the same strategy to determine whether the ATRX–Daxx complex becomes smaller in an extract from a cell line derived from a patient with ATRX syndrome that contains ATRX at a very low level (Fig. 2d). So far, all ATRX patient cell lines have a residual level of full-length ATRX, even though nonsense or frameshift mutations were found near the N terminus of ATRX gene products. The patient cells apparently have some read-through of premature termination codons (R.G., unpublished data).
The level of Daxx in the nuclear extract of the ATRX patient cell line is comparable to that from HeLa cells (Fig. 2d). After Superose 6 fractionation, the expected Daxx peak corresponding the ATRX–Daxx complex (Fig. 2c, fraction 20) is almost gone; only the second, “non-ATRX” peak of Daxx remains (around fraction 25). The residual ATRX in this cell line still peaked in fraction 20. These results confirmed that the 1-MDa complex corresponds to that of ATRX–Daxx, whereas the 700-kDa Daxx complex is independent of ATRX.
ATRX Complex Alters the DNase I Digestion of a Nucleosome in the Presence of ATP. A mononucleosome disruption assay (7, 20, 21) was used to determine whether ATRX complex has chromatin-remodeling activity. In this assay, a 176-bp fragment of DNA containing a nucleosome positioning sequence from sea urchin 5S rRNA gene was assembled into a rotationally phased mononucleosome. DNase I digestion of this nucleosome produces a pattern of distinctive 10-bp ladders on denaturing PAGE (Fig. 3a, lane 2 vs. 1). The human SWI/SNF complex disrupted the 10-bp ladders in the presence of ATP but not with nonhydrolyzable ATP-γ-S (Fig. 3a, lanes 5–7). This can be inferred because those nucleotides inaccessible to DNase I in the unremodeled nucleosome now become accessible, suggesting that these nucleotides changed their phasing after chromatin remodeling. This finding is consistent with a model that the DNA phasing of these nucleosomes has been randomized by SWI/SNF (39).
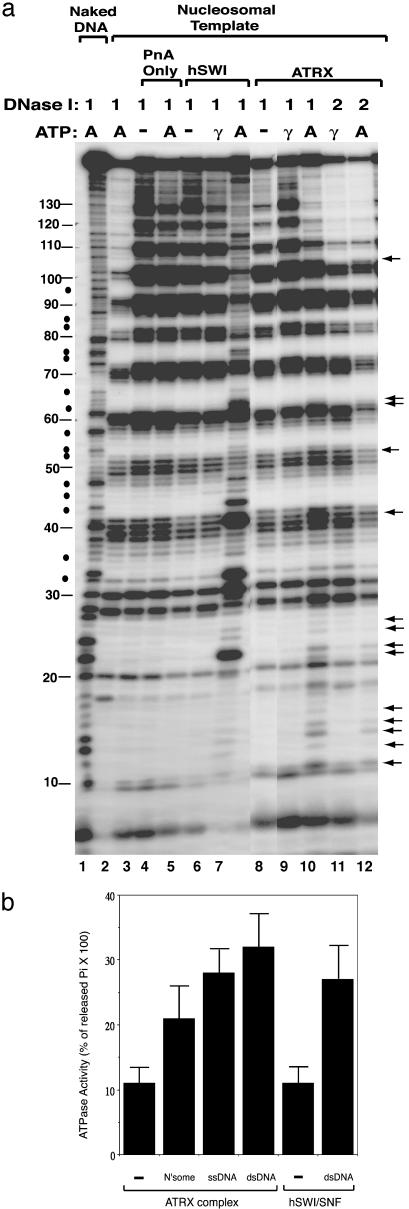
Fig. 3.
The ATRX complex alters the DNase I digestion pattern of a nucleosome in the presence of ATP. (a) Autoradiograph showing the results of the mononucleosome disruption assay. Complexes isolated by protein A alone (PnA) or Abs against ATRX and human SWI/SNF (hSWI) are indicated at the top. The templates used and the presence of ATP (A) or ATP-γ-S (γ) are indicated. The amounts of DNase used, 1 and 2, represent 0.2 and 0.4 units, respectively. The solid arrows mark the nucleotides whose digestion was enhanced by the ATRX–Daxx complex in the presence of ATP. The solid dots denote the nucleotides between the 10-bp ladders whose digestion was stimulated by human SWI/SNF but not ATRX–Daxx. The solid lines mark the positions of the nucleotides in 5S DNA. The 10-bp ladders can be observed between nucleotides 10 and 130 of 5S DNA. (b) Graphic presentation showing that the ATRX complex has DNA- or nucleosome-stimulated ATPase activity (shown in percentage of released inorganic phosphate from total ATP multiplied by 100). The ATPase activity of mock immunoprecipitation (using preimmune serum) is indistinguishable from that of the background (using protein A beads), which was subtracted during calculation. The presence of nucleosomes (N′some), single-stranded DNA (ssDNA), and double-stranded DNA (dsDNA) is indicated.
ATRX complex had no obvious effect on the DNase I digestion pattern in the absence of ATP or in the presence of ATP-γ-S (Fig. 3a, lanes 8, 9, and 11), but in the presence of ATP, the complex altered the digestion pattern (Fig. 3a, lanes 10 and 12). The region that shows the most alteration is between nucleotides 10 and 30 of the 5S DNA where nucleotides facing inside toward the nucleosome showed an increased level of digestion. This region is near the entry site of the nucleosome, because nucleotides before 10 are accessible to DNase I digestion. For regions beyond the first 30 nucleotides of 5S DNA, only certain nucleotides facing out from the nucleosome exhibited enhanced digestion, whereas nucleotides facing in show no enhancement. The results suggest that the ATRX complex mainly disrupts the DNA–histone interaction at the entry site of the nucleosome, and disruption does not alter nucleosome phasing. This is different from remodeling by SWI/SNF (compare Fig. 3a, lanes 10 and 7).
The ATRX complex was found to have an ATPase activity that can be stimulated by DNA or nucleosomes (Fig. 3b), a property that resembles SWI/SNF. Different preparations of ATRX complex with ATPase activity 1- to 2-fold that of SWI/SNF have been used for the mononucleosome disruption assay, with similar results obtained.
The ATRX–Daxx Complex Has an ATP-Dependent Triple-Helix Displacement Activity. The yeast RSC and SWI/SNF chromatin-remodeling complexes, as well as the ATPase subunit of RSC, STH1p, have been shown to be ATP-dependent DNA translocases (23, 40). One way to show translocation is the triple-helix displacement assay. In this assay, a triple helix (H-DNA) is formed in which each nucleotide of the third strand forms Hoogsteen base pairs with Watson–Crick base pairs of the duplex (41). When the translocase proceeds through the triple helix, the third strand is displaced. Complexes isolated by different ATRX and Daxx Abs all displayed triple-helix displacement activity (Fig. 4a), suggesting that the ATRX–Daxx complex has a translocase property like yeast RSC and SWI/SNF.
One possible mechanism for the ATRX–Daxx complex to displace triple helix is through direct binding and dissociation. However, complexes isolated by either ATRX or Daxx do not displace the third strand in a blunt triplex (Fig. 4c). This result is consistent with the idea that the ATRX–Daxx complex needs to bind the flanking duplex DNA and translocate into the triple helix to displace the third strand.
We also investigated whether ATRX–Daxx may function as a DNA helicase. Using a standard duplex DNA displacement assay, complexes isolated by either ATRX or Daxx Ab showed no detectable activity (Fig. 4d), consistent with that ATRX–Daxx is functioning as a chromatin-remodeling complex but not a helicase.
Daxx Is Dispensible for the Triple-Helix Displacement Activity of ATRX Complex. We examined whether Daxx is required for the triple-helix displacement activity of the ATRX–Daxx complex. Daxx was completely washed away from ATRX by using buffer containing salt of 0.5 M or higher (Fig. 4e; silver-staining gel not shown). The remaining ATRX protein displays triplex activity indistinguishable from that of the ATRX–Daxx complex (Fig. 4f), suggesting that Daxx is probably not needed for the remodeling activity of ATRX.
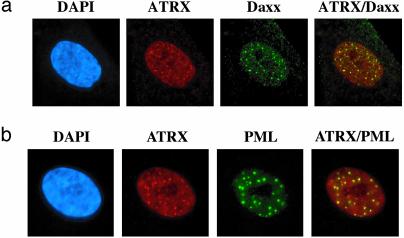
ATRX and Daxx Colocalize in PML Nuclear Bodies. ATRX is predominantly associated with heterochromatin both in interphase and metaphase (12). However, a significant proportion of ATRX is present in nuclear speckles in human cells (17). The distribution of ATRX between heterochromatin and nuclear speckles varies from one cell type to another. Consistent with previous observations (12, 17), ATRX was found to localize in the nuclei of interphase human cells with a typical speckled pattern (Fig. 5). Daxx was found to display a similar speckled pattern in the nuclei, which is also in agreement with prior studies (28–30, 32, 33). Importantly, the majority of ATRX signals colocalize with those of Daxx, providing a further indication that these two proteins may work together in vivo.
Fig. 5.
ATRX and Daxx colocalize in PML nuclear bodies. (a) Immunofluorescence images showing colocalization of ATRX and Daxx in human fibroblast cell lines. Images for each protein and the merged image are shown. (b) Immunofluorescence images showing colocalization of ATRX with PML in human fibroblasts. The nucleus was costained with 4′,6-diamidino-2-phenylindole (DAPI).
Daxx has been shown to associate with the PML gene product, PML, and localize in PML nuclear bodies (29, 30, 32, 33). PML frequently is involved in chromosomal translocations in acute PML. In normal cells, PML is concentrated within 10–20 nuclear structures known as PML bodies, which are believed to play roles in transcriptional activation, DNA replication, apoptosis, and viral infection. The colocalization of ATRX and Daxx raised the possibility that ATRX also localizes in PML bodies. Costaining experiments showed that the majority of ATRX foci colocalize with those of PML (Fig. 5c), suggesting that the ATRX–Daxx complex may play a role in the function of PML bodies.
Discussion
In this study, we show that ATRX, the protein involved in ATRX syndrome, constitutes a novel complex with Daxx. This complex has several activities consistent with it being a chromatin-remodeling complex. They include ATP-dependent activity that alters the DNase I digestion pattern of a nucleosome and ATP-dependent triple-helix displacement activity. These data support the notion that ATRX functions as part of a chromatin-remodeling complex and may thereby participate in transcriptional regulation like other remodeling complexes. In addition, we found that the ATRX–Daxx complex lacks any detectable DNA helicase activity, distinguishing it from proteins involved in DNA damage repair and genome instability diseases (e.g., Bloom syndrome and Werner syndrome). This finding is consistent with previously published clinical data that ATRX patients have no increased incidence of chromosome abnormalities or increased propensity to develop malignancy.
Interestingly, the ATRX–Daxx complex does not randomize DNA phasing of a nucleosome, which is in contrast to remodeling by SWI/SNF or NURD complexes. Its disruption of the DNase I digestion pattern of a mononucleosome occurs mainly near the entry site of the nucleosome, which is also different from remodeling by SWI/SNF or NURD. The ATPase motif of ATRX is most similar to that in Rad54 (42). Rad54 displays an ATP-dependent chromatin-remodeling activity in the presence of Rad51 (43). A recent study has shown that Rad54 has remodeling properties similar to ATRX, including triplex displacement activity and failure to randomize DNA phasing of nucleosomes (40). Speculatively, ATRX–Daxx and Rad54 may be members of a group of complexes that remodel chromatin differently from the SWI/SNF and NURD families.
How may ATRX and Daxx work together in a complex? Chromatin-remodeling complexes are often recruited to their targets by sequence-specific transcription factors that bind to these promoters (44). Several lines of evidence hint that Daxx may be a targeting subunit for ATRX complex. First, Daxx contains two regions predicted to form paired amphipathic helices that resemble those of Sin3 (31). A paired amphipathic helix motif in Sin3 targets the Sin3 complex through interactions with a sequence-specific transcription factor (45). The paired amphipathic helix regions of Daxx may play a targeting role analogous to that of Sin3. Second, by yeast two-hybrid screens, Daxx interacts with several sequence-specific transcription factors (31, 34, 35). Third, in ongoing work, we found that the two other major Daxx-associated polypeptides, Daxx-associated polypeptide-50 and -60 (Fig. 2a), are both sequence-specific transcription factors (unpublished data). Our data are therefore consistent with a model that the ATRX–Daxx complex participates in chromatin remodeling for genes that are controlled by Daxx-interacting sequence-specific transcription factors. The molecular defects in ATRX patients may result from inappropriate regulation of these target genes.
Acknowledgments
We thank Drs. B. Cairns, C. Peterson, A. Saha, J. Cote, T. Owen-Hughes, and C. Wu for reagents and advice on different assays; Drs. H. Lu, D. Reinberg, M. Baron, and S. Lippard for Abs; Dr. N. Sherman for MS analysis; and Drs. C. Peterson and Y. Zhang for communicating unpublished results. We thank the National Cell Culture Center for providing cells. We also thank Dr. D. Schlessinger for critical reading of the manuscript. W.W. is a scholar from The Ellison Medical Foundation and has received a grant from the Rett Syndrome Foundation.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviation: PML, promyelocytic leukemia.
References
- 1.Krebs, J. E. & Peterson, C. L. (2000) Crit. Rev. Eukaryotic Gene Expression 10, 1–12. [PubMed] [Google Scholar]
- 2.Neely, K. & Workman, J. (2002) Biochim. Biophys. Acta 1603, 19–29. [DOI] [PubMed] [Google Scholar]
- 3.Kwon, H., Imbalzano, A. N., Khavari, P. A., Kingston, R. E. & Green, M. R. (1994) Nature 370, 477–481. [DOI] [PubMed] [Google Scholar]
- 4.Wang, W., Cote, J., Xue, Y., Zhou, S., Khavari, P. A., Biggar, S. R., Muchardt, C., Kalpana, G. V., Goff, S. P., Yaniv, M., et al. (1996) EMBO J. 15, 5370–5382. [PMC free article] [PubMed] [Google Scholar]
- 5.Xue, Y., Canman, J. C., Lee, C. S., Nie, Z., Yang, D., Moreno, G. T., Young, M. K., Salmon, E. D. & Wang, W. (2000) Proc. Natl. Acad. Sci. USA 97, 13015–13020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lemon, B., Inouye, C., King, D. S. & Tjian, R. (2001) Nature 414, 924–938. [DOI] [PubMed] [Google Scholar]
- 7.Xue, Y., Wong, J., Moreno, G. T., Young, M. K., Cote, J. & Wang, W. (1998) Mol. Cell 2, 851–861. [DOI] [PubMed] [Google Scholar]
- 8.Gibbons, R. J., Picketts, D. J., Villard, L. & Higgs, D. R. (1995) Cell 80, 837–845. [DOI] [PubMed] [Google Scholar]
- 9.Gibbons, R. J. & Higgs, D. R. (2000) Am. J. Med. Genet. 97, 204–212. [DOI] [PubMed] [Google Scholar]
- 10.Gibbons, R. J., Bachoo, S., Picketts, D. J., Aftimos, S., Asenbauer, B., Bergoffen, J., Berry, S. A., Dahl, N., Fryer, A., Keppler, K., et al. (1997) Nat. Genet. 17, 146–148. [DOI] [PubMed] [Google Scholar]
- 11.Berube, N. G., Jagla, M., Smeenk, C., De Repentigny, Y., Kothary, R. & Picketts, D. J. (2002) Hum. Mol. Genet. 11, 253–261. [DOI] [PubMed] [Google Scholar]
- 12.McDowell, T. L., Gibbons, R. J., Sutherland, H., O'Rourke, D. M., Bickmore, W. A., Pombo, A., Turley, H., Gatter, K., Picketts, D. J., Buckle, V. J., et al. (1999) Proc. Natl. Acad. Sci. USA 96, 13983–13988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cardoso, C., Timsit, S., Villard, L., Khrestchatisky, M., Fontes, M. & Colleaux, L. (1998) Hum. Mol. Genet. 7, 679–684. [DOI] [PubMed] [Google Scholar]
- 14.Le Douarin, B., Nielsen, A. L., Garnier, J. M., Ichinose, H., Jeanmougin, F., Losson, R. & Chambon, P. (1996) EMBO J. 15, 6701–6715. [PMC free article] [PubMed] [Google Scholar]
- 15.Gibbons, R. J., McDowell, T. L., Raman, S., O'Rourke, D. M., Garrick, D., Ayyub, H. & Higgs, D. R. (2000) Nat. Genet. 24, 368–371. [DOI] [PubMed] [Google Scholar]
- 16.Guerrini, R., Shanahan, J. L., Carrozzo, R., Bonanni, P., Higgs, D. R. & Gibbons, R. J. (2000) Ann. Neurol. 47, 117–121. [PubMed] [Google Scholar]
- 17.Berube, N. G., Smeenk, C. A. & Picketts, D. J. (2000) Hum. Mol. Genet. 9, 539–547. [DOI] [PubMed] [Google Scholar]
- 18.Meetei, A. R., Sechi, S., Wallisch, M., Yang, D., Young, M. K., Joenje, H., Hoatlin, M. E. & Wang, W. (2003) Mol. Cell. Biol. 23, 3417–3426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang, W., Xue, Y., Zhou, S., Kuo, A., Cairns, B. R. & Crabtree, G. R. (1996) Genes Dev. 10, 2117–2130. [DOI] [PubMed] [Google Scholar]
- 20.Owen-Hughes, T., Utley, R. T., Cote, J., Peterson, C. L. & Workman, J. L. (1996) Science 273, 513–516. [DOI] [PubMed] [Google Scholar]
- 21.Wang, W., Chi, T., Xue, Y., Zhou, S., Kuo, A. & Crabtree, G. R. (1998) Proc. Natl. Acad. Sci. USA 95, 492–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao, K., Wang, W., Rando, O. J., Xue, Y., Swiderek, K., Kuo, A. & Crabtree, G. R. (1998) Cell 95, 625–636. [DOI] [PubMed] [Google Scholar]
- 23.Saha, A., Wittmeyer, J. & Cairns, B. R. (2002) Genes Dev. 16, 2120–2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shen, J. C., Gray, M. D., Oshima, J., Kamath-Loeb, A. S., Fry, M. & Loeb, L. A. (1998) J. Biol. Chem. 273, 34139–34144. [DOI] [PubMed] [Google Scholar]
- 25.Michaelson, J. S. (2000) Apoptosis 5, 217–220. [DOI] [PubMed] [Google Scholar]
- 26.Lai, J. S. & Herr, W. (1992) Proc. Natl. Acad. Sci. USA 89, 6958–6692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang, X., Khosravi-Far, R., Chang, H. Y. & Baltimore, D. (1997) Cell 89, 1067–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pluta, A. F., Earnshaw, W. C. & Goldberg, I. G. (1998) J. Cell Sci. 111, 2029–2041. [DOI] [PubMed] [Google Scholar]
- 29.Torii, S., Egan, D. A., Evans, R. A. & Reed, J. C. (1999) EMBO J. 18, 6037–6049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhong, S., Salomoni, P., Ronchetti, S., Guo, A., Ruggero, D. & Pandolfi, P. P. (2000) J. Exp. Med. 191, 631–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hollenbach, A. D., Sublett, J. E., McPherson, C. J. & Grosveld, G. (1999) EMBO J. 18, 3702–3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ishov, A. M., Sotnikov, A. G., Negorev, D., Vladimirova, O. V., Neff, N., Kamitani, T., Yeh, E. T., Strauss, J. F., III, & Maul, G. G. (1999) J. Cell Biol. 147, 221–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li, H., Leo, C., Zhu, J., Wu, X., O'Neil, J., Park, E. J. & Chen, J. D. (2000) Mol. Cell. Biol. 20, 1784–1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li, R., Pei, H., Watson, D. K. & Papas, T. S. (2000) Oncogene 19, 745–753. [DOI] [PubMed] [Google Scholar]
- 35.Emelyanov, A. V., Kovac, C. R., Sepulveda, M. A. & Birshtein, B. K. (2002) J. Biol. Chem. 277, 11156–11164. [DOI] [PubMed] [Google Scholar]
- 36.Michaelson, J. S., Bader, D., Kuo, F., Kozak, C. & Leder, P. (1999) Genes Dev. 13, 1918–1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hollenbach, A. D., McPherson, C. J., Mientjes, E. J., Iyengar, R. & Grosveld, G. (2002) J. Cell Sci. 115, 3319–3330. [DOI] [PubMed] [Google Scholar]
- 38.Peterson, C. L., Dingwall, A. & Scott, M. P. (1994) Proc. Natl. Acad. Sci. USA 91, 2905–2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cote, J., Peterson, C. L. & Workman, J. L. (1998) Proc. Natl. Acad. Sci. USA 95, 4947–4952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jaskelioff, M., Van Komen, S., Krebs, J. E., Sung, P. & Peterson, C. L. (2003) J. Biol. Chem. 278, 9212–9218. [DOI] [PubMed] [Google Scholar]
- 41.van Dongen, M. J., Doreleijers, J. F., van der Marel, G. A., van Boom, J. H., Hilbers, C. W. & Wijmenga, S. S. (1999) Nat. Struct. Biol. 6, 854–859. [DOI] [PubMed] [Google Scholar]
- 42.Picketts, D. J., Tastan, A. O., Higgs, D. R. & Gibbons, R. J. (1998) Mamm. Genome 9, 400–403. [DOI] [PubMed] [Google Scholar]
- 43.Alexiadis, V. & Kadonaga, J. T. (2002) Genes Dev. 16, 2767–2771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hassan, A. H., Neely, K. E., Vignali, M., Reese, J. C. & Workman, J. L. (2001) Front. Biosci. 6, D1054–D1064. [DOI] [PubMed] [Google Scholar]
- 45.Brubaker, K., Cowley, S. M., Huang, K., Loo, L., Yochum, G. S., Ayer, D. E., Eisenman, R. N. & Radhakrishnan, I. (2000) Cell 103, 655–665. [DOI] [PubMed] [Google Scholar]