Abstract
An enantio-specific polyphenol oxidase (PPO) was purified ≈1,700-fold to apparent homogeneity from the creosote bush (Larrea tridentata), and its encoding gene was cloned. The posttranslationally processed PPO (≈43 kDa) has a central role in the biosynthesis of the creosote bush 8–8′ linked lignans, whose representatives, such as nordihydroguaiaretic acid and its congeners, have potent antiviral, anticancer, and antioxidant properties. The PPO primarily engenders the enantio-specific conversion of (+)-larreatricin into (+)-3′-hydroxylarreatricin, with the regiochemistry of catalysis being unambiguously established by different NMR spectroscopic analyses; the corresponding (–)-enantiomer did not serve as a substrate. This enantio-specificity for a PPO, a representative of a widespread class of enzymes, provides additional insight into their actual physiological roles that hitherto have been difficult to determine.
In the traditional medicine of the indigenous peoples of North and South America, the creosote bush (Larrea tridentata) was widely used for treatment of various ailments including digestive disorders, rheumatism, venereal diseases, and sores (1–3). The main bioactive constituents apparently reside in the resinous exudate (≈5–10% of leaf dry weight), the major constituent of which is the powerful antioxidant nordihydroguaiaretic acid (NDGA, 10, Fig. 1) (4, 5). The latter is used mainly in nonfood applications, including as a polymer (e.g., rubber) stabilizer, an antioxidant in perfumery oil and photographic formulations (5, 6), and antihyperglycemic (7) and skin anti-aging agents (8). NDGA (10) also has allelopathic properties and inhibits growth of encroaching plant species that compete for water and nutrients (9).
Fig. 1.
Proposed biosynthetic pathway to creosote bush (L. tridentata) lignans.
More recently, anticancer (10) and antiviral (11, 12) properties were reported for NDGA (10) and its derivatives: e.g., 3′-O-methyl-NDGA (11) has anti-HIV activities by inhibiting Tat-regulated HIV transactivation (11), whereas other methylated/acetylated NDGA analogues display promising antiviral activities against HIV, herpes simplex, and human papilloma (12).
NDGA (10) and its various structural analogues in L. tridentata (2–9, 11–14, see Fig. 1) (2, 13–16) belong to a class of plant natural products known as lignans, these being frequently found as optically active dimers (17, 18). Whereas all known L. tridentata lignans are linked via 8–8′ bonds, they exist in dibenzyltetrahydrofuran, dibenzylbutane, and aryltetrahydronaphthalene forms (16). Based on structural considerations, all appear to be allylphenol (e.g., anol 1) derived (16), rather than the more common lignan skeleta in other plants, which are of monolignol (e.g., coniferyl alcohol 15) origin (18). However, nothing is yet conclusively known about allylphenol coupling (in L. tridentata and other species) and/or subsequent postcoupling transformations, such as aromatic ring hydroxylations, etc. Allylphenol-derived lignan formation is in contrast to entry into the monolignol-derived lignans, which can occur via stereo-selective coupling of E-coniferyl alcohol (15, Scheme 1) to yield, for example, (+)-pinoresinol (16) in Forsythia intermedia; the latter conversion involves participation of a dirigent (Latin; dirigere, to guide or to align) protein and an oxidase (19–21).
Scheme 1.
In this investigation, the biosynthetic pathway to the antiviral lignans in the creosote bush was initially examined by means of radiochemical precursor administration experiments together with metabolite analysis and identification. Of the various 8–8′ regio-specifically coupled diastereomers formed in vivo, (+)-larreatricin (2) underwent enantiospecific hydroxylation primarily to give (+)-3′-hydroxylarreatricin (6), whereas the (–)-form of 2 was not metabolized. This enantio-specific conversion was catalyzed by a polyphenol oxidase (PPO), whose encoding gene was cloned. Because (+)-3′-hydroxylarreatricin (6) is a presumed precursor of the optically inactive meso lignan, NDGA (10), these findings now provide needed insight into how various antiviral and anticancer creosote bush lignans are formed in vivo, including the nature of their regio- and enantio-specific conversions.
This enantio-specificity is also of particular interest, as the precise physiological roles of PPOs have hitherto been largely difficult to ascertain, because they can act on a wide variety of aromatic substrates (22–25), which has led to significant uncertainty about their actual roles in vivo.
Materials and Methods
Materials, Instrumentation, and Chemical Syntheses. See Supporting Methods and Tables 2–4, which are published as supporting information on the PNAS web site, www.pnas.org.
Chiral HPLC Analyses. Synthetic (±)-larreatricins (2) were individually separated into their enantiomeric forms by chiral HPLC as described (16), whereas (±)-3′-hydroxylarreatricins (6) were resolved by using a Chirobiotic V column (250 × 4.6 mm inner diameter, Advanced Separation Technologies, Whippany, NJ), eluted with ethanol/hexanes (1:4) at a flow rate of 1 ml·min–1, with detection both at 280 nm and by an in-line laser Advanced Laser Polarimeter (cell volume 56 mm3, length 5.17 cm, PDR-Chiral, Palm Beach Gardens, FL) connected in series (16, 26).
Metabolism of l-[U-14C]Phe. Ten 2-month-old L. tridentata seedlings were individually placed in microcentrifuge tubes (1.5 ml), with each containing a solution of l-[U-14C]Phe in H2O (185 kBq, 100 μl). After l-[U-14C]Phe uptake over ≈1 h, the seedlings were placed in a growth chamber (27°C day/22°C night with a photoperiod of 16 h) for different time periods (1–6 h) with H2O replenished as needed. For each selected time period, individual seedlings were harvested, frozen (liquid N2), and ground to a powder in a mortar by means of a pestle, with each sample extracted with MeOH/EtOAc (1:1, 7 ml). The MeOH/EtOAc extract from each plant sample was then evaporated to dryness in vacuo, with the individual residues dissolved in MeOH/H2O (1:1, 1 ml). An aliquot (100 μl) of each was next subjected to RP-HPLC analysis (Nova-Pak C18 column, Waters) eluted as follows with detection at 280 nm: flow rate of 1 ml·min–1; isocratic solvent system A/B (CH3CN/3% HOAc in H2O) (28:72) for the first 10 min followed by a linear A/B gradient from 28:72 to 60:40 between 10 and 40 min. Fractions were collected at 1-min intervals and subjected to radiochemical scintillation counting analysis. To identify the lignans corresponding to the radioactive peaks, L. tridentata seedlings (10 g) were extracted with MeOH/EtOAc (1:1, 30 ml), and the resulting extract was evaporated to dryness in vacuo. The MeOH/EtOAc residue was redissolved in MeOH/H2O (1:1, 1.2 ml), and multiple aliquots (40 μl) were sequentially subjected to RP-HPLC using the elution conditions described above. Peaks corresponding to previously isolated radioactive components were individually collected and freeze-dried. Each compound obtained was further purified by RP-HPLC and identified by both 1H NMR and electron impact-MS analyses. As an example, radiolabel incorporations into the various L. tridentata lignans after 1- and 6-h metabolism, respectively, of l-[U-14C]Phe were as follows: 10 (0.2% and 0.6%), 11 (0.06% and 0.1%), 12 (0.03% and 0.1%), 13 (0.1% and 0.2%), and 14 (0.05% and 0.1%).
Enzymatic Formation of 3′-Hydroxylarreatricin (6). Each assay consisted of (±)-larreatricins (2), ascorbic acid (10 mM), and 3′-larreatricin hydroxylase preparations (obtained at various stages of purification) in potassium phosphate buffer (50 mM, pH 7.0) to a total volume of 100 μl. Assays were initiated by addition of enzyme preparation, and after 1-h incubation at 30°C with shaking, each enzymatic reaction mixture was extracted twice with EtOAc (500 μl). [Controls were carried out in the absence of ascorbic acid.] After centrifugation (13,800 × g, 5 min), the EtOAc solubles were removed and evaporated to dryness in vacuo, then reconstituted in MeOH/H2O (1:1, 120 μl) with an aliquot (100 μl) subjected to RP-HPLC analysis (SymmetryShield RP18 column, Waters) as described for racemic 6 and 7 in Supporting Methods; 3′-hydroxylarreatricin (6) amounts formed were quantified by using a previously established standard curve.
To identify the reaction products, a large-scale enzymatic incubation was carried out with (±)-larreatricins (2) (9.1 mg) in 8 ml of assay mixture (as described above) and extracted with EtOAc (3 × 6 ml). The EtOAc solubles were combined and evaporated to dryness in vacuo, reconstituted in MeOH/H2O (1:1, 1.4 ml), with the enzymatically formed 6 (and 7) purified by HPLC as described above.
(+)-3′-hydroxylarreatricin (6) characterization is as follows. 1H NMR (500 MHz, DMSO-d6): δ 0.49 (3H, d, J9′,8′ = 7.1 Hz, 9′-3H), 0.86 (3H, d, J9,8 = 6.7 Hz, 9-3H), 2.30 (1H, m, J8,7 = 9.5, J8,8′ = 5.9, J8,9 = 6.7 Hz, 8-H), 2.35 (1H, m, J8′,7′ = 4.4, J8′,8 = 5.9, J8′,9′ = 7.1 Hz, 8′-H), 4.47 (1H, d, J7,8 = 9.5 Hz, 7-H), 5.24 (1H, d, J7′,8′ = 4.4 Hz, 7′-H), 6.52 (1H, dd, J6′,5′ = 8.0, J6′,2′ = 1.7 Hz, 6′-H), 6.66 (1H, d, J5′,6′ = 8.0 Hz, 5′-H), 6.68 (1H, d, J2′,6′ = 1.7 Hz, 2′-H), 6.72 (2H, d, J3,5 = 8.5 Hz, 3-H, 5-H), 7.16 (2H, d, J2,6 = 8.5 Hz, 2-H, 6-H), 8.66 (1H, s, 4′-OH), 8.75 (1H, s, 3′-OH), 9.30 (1H, s, 4-OH). 13C NMR (125.67 MHz, DMSO-d6): δ 9.3 (9′-C), 11.6 (9-C), 42.4 (8′-C), 46.7 (8-C), 83.7 (7′-C), 84.6 (7-C), 113.4 (2′-C), 114.9 (3-C, 5-C), 115.1 (5′-C), 116.6 (6′-C), 127.4 (2-C, 6-C), 131.6 (1′-C), 133.4 (1-C), 143.7 (4′-C), 144.7 (3′-C), 156.6 (4-C). For 1H, 13C, heteronuclear multibond connectivity (HMBC), and nuclear Overhauser effect (NOE) spectroscopic assignments see Table 4. Electrospray ionization–Fourier transform MS (m/z): [M–H], found 299.1291; [C18H20O4–H] requires 299.1288; [M + Cl], found 335.1050; [C18H20O4 + Cl] requires 335.1055.
Purification of (+)-Larreatricin 3′-Hydroxylase. Protein purification procedures were carried out at 4°C unless specified otherwise, with chromatographic eluants monitored at 280 nm; protein concentrations were determined by using the method of Bradford (27). (+)-Larreatricin 3′-hydroxylase was purified from 5 kg of L. tridentata twigs; however, because of the nature of the tissue used only 100 g of tissue was processed at a time as described below.
L. tridentata 3-month-old twigs (100 g) were frozen in liquid N2 and pulverized by using a Waring blender, with the homogenate transferred to a mortar. After further grinding to a fine powder, polyvinylpolypyrrolidone (10 g) was added with the resulting powder homogenized in Tris·HCl buffer (0.1 M, pH 7.5, 200 ml) containing 250 mM sucrose, 50 mM NaCl, 5 mM DTT, and 0.1 mM PMSF. The whole was centrifuged (20,000 × g) for 30 min, with the supernatant filtered through Mira-cloth and further subjected to ultracentrifugation (160,000 × g, 90 min). The pellet was homogenized in Tris·HCl buffer (0.1 M, pH 7.5, 5 ml) containing Triton X-100 (2%, vol/vol) with a glass-glass homogenizer, with the homogenate subjected to ultracentrifugation (100,000 × g, 60 min) to give a supernatant (designated as protein preparation), which was stored at –80°C until needed. This procedure was repeated 50 times, yielding a total of 9.27 g of protein (Table 1).
Table 1. Purification of (+)-larreatricin 3′-hydroxylase.
| Purification step | Protein, mg | Total activity, nmol·h-1 | Specific activity, nmol·h-1·mg-1 | Purification factor, fold | Yield, % |
|---|---|---|---|---|---|
| Membrane proteins | 9,265 | 735,670 | 79.40 | — | 100 |
| DEAE-Sepharose | 98.44 | 197,470 | 2,006 | 25 | 26.8 |
| Hydroxylapatite | 1.87 | 149,950 | 89,018 | 1,120 | 20.4 |
| Phenyl-Sepharose | 0.42 | 55,120 | 133,180 | 1,680 | 7.5 |
Protein preparations (1.85 g protein total) obtained from L. tridentata twigs (100 g × 10 preparations) were combined and loaded onto a Sephadex G-25 column (2.5 i.d. × 18 cm) equilibrated in buffer A (potassium phosphate buffer, 50 mM, pH 7.0), with the eluted protein fraction applied to a DEAE Sepharose column (2.6 i.d. × 15 cm) previously equilibrated in buffer A. After washing with buffer A (80 ml), proteins were desorbed with a linear gradient of 0 to 1 M NaCl in buffer A (800 ml). Fractions containing larreatricin 3′-hydroxylase activity, eluting between 650 and 880 mM NaCl, were combined, concentrated to ≈7.5 ml, then desalted on a PD10 column previously equilibrated and eluted in buffer B (potassium phosphate buffer, 10 mM, pH 7.0) and stored at –80°C until required. [These chromatographic steps were repeated four times with previously combined protein preparations from L. tridentata twigs (100 g × 10, each).]
Active fractions from the DEAE columns and desalted on PD10 columns (98.44 mg protein total) were thawed, combined, and next applied to a hydroxylapatite column (1.6 mm i.d. × 18 cm) equilibrated in buffer B. After washing with buffer B (40 ml), proteins were eluted with a linear gradient from 10 to 800 mM potassium phosphate buffer (pH 7.0, 400 ml). Fractions containing larreatricin 3′-hydroxylase activity, eluting between 550 and 750 mM potassium phosphate buffer, were combined, concentrated to ≈5 ml, and desalted on a PD-10 column equilibrated in buffer A. The desalted fraction (1.87 mg protein) was brought to a final concentration of 1 M (NH4)2SO4 and further applied to a Phenyl Sepharose column (1 i.d. × 7 cm) equilibrated in buffer C [potassium phosphate buffer, 50 mM, pH 7.0 containing 1 M (NH4)2SO4]. After washing with buffer C (20 ml), proteins were desorbed by using a linear gradient from 100% buffer C to 100% buffer A in 100 ml. Fractions with larreatricin 3′-hydroxylase activity, eluting between 500 and 900 mM (NH4)2SO4, were combined, concentrated to ≈5 ml (0.42 mg protein), and stored at –80°C until required.
Peptide Sequencing. The partially purified larreatricin 3′-hydroxylase obtained after Phenyl Sepharose column chromatography was subjected to SDS/PAGE (4–15% Ready gel, Bio-Rad), with proteins visualized by using a Colloidal blue staining kit (Invitrogen). The major band, corresponding to (+)-larreatricin 3′-hydroxylase, was excised from the gel, with the section washed with 50% aqueous CH3CN (vol/vol), this being (subsequently) subjected to trypsin digestion and sequenced at the Harvard Microchemistry Facility (Cambridge, MA) by microcapillary RP-HPLC nano-electrospray tandem MS (μLC/MS/MS) on a Finnigan LCQ DECA XP quadrupole ion trap mass spectrometer; the MS/MS data so obtained were correlated with known amino acid sequences in various protein databases (28–30). The sequences of two tryptic fragments were found to have very high homology to the conserved copper binding domains A and B in Vicia faba PPO (WYLYFYER and DPIFYSHHSNVDR, GenBank accession no. Z11702) (31) and to the copper binding domain B from apricot (Prunus armeniaca) PPO, (DPLFYAHHANVDR, GenBank accession no. AF020786) (32), respectively.
Construction of L. tridentata Leaf cDNA Library and Cloning of the cDNA Encoding (+)-Larreatricin 3′-Hydroxylase. See Supporting Methods.
Results and Discussion
On a structural basis, the simplest L. tridentata lignans are larreatricin (2), 3,3′ didemethoxyverrucosin (3), meso-3,3′ didemethoxynectandrin B (4), and 8′-epi-larreatricin (5), with all apparently being directly E-p-anol (1) derived (16). Chiral HPLC analysis further revealed that both larreatricin (2) and 8′-epilarreatricin (5) were present in ≈92% and 98% enantiomeric excess, respectively, as the corresponding (–)-antipodes, whereas didemethoxyverrucosin (3) and didemethoxynectandrin B (4) were in near racemic and meso forms, respectively. No other creosote bush lignans with different interunit linkages (e.g., 8–3′,8–O–4′, etc.) were detected in either this or any other study (14–16). Accordingly, these data provide strong evidence for regio-specific 8–8′ coupling control, but give no insight into whether formation of larreatricin (2) and 8′ epi-larreatricin (5) either involves stereo-selective coupling, or if one of the antipodes is differentially metabolized after regio-specific 8–8′ coupling.
In this study, l-[U-14C]Phe was first administered to creosote bush seedlings over different time intervals (1–6 h) to establish whether the lignans were phenylpropanoid pathway metabolites and to examine the pattern of radiolabeling in each. This precursor was primarily metabolized into [14C]-NDGA (10), [14C]-3-O-methyl NDGA (11), [14C]-dihydroguaiaretic acid (12), [14C]-3,3′-di-O-demethylisoguaiacin (13), [14C]-norisoguaiacin (14), and to a lesser extent into [14C]-larreatricin (2), [14C]-8′-epi-larreatricin (5) and [14C]-3′-hydroxy-8′-epi larreatricin (8), with highest incorporation levels being noted in NDGA (10) even after 1-h metabolism (see Materials and Methods).
From these data, together with the known lignan skeletal types present in the creosote bush (L. tridentata), a tentative biosynthetic scheme to the most abundant lignans such as meso (optically inactive) NDGA (10) and its analogs can be proposed (Fig. 1), i.e., where Phe is metabolized into E-p-anol (1), this in turn undergoing (at minimum) regio-specific coupling to give the furanolignans 2–5. However, given that the most abundant dibenzylbutane (e.g., 10–12) and aryltetrahydronaphthalene (e.g., 13 and 14) lignans from L. tridentata have the same configurations at C-8/C-8′ as that of larreatricin (2), the latter can be envisaged to serve as a central intermediate in their biosynthetic pathways. [By contrast, 8′-epi-larreatricin (5) is apparently only hydroxylated to generate the presumed end product, 3′-hydroxy-8′-epi-larreatricin (8), whereas 3,3′-didemethoxyverrucosin (3) and 3,3′-didemethoxynectandrin B (4) do not appear to be metabolized further.]
It was thus deduced that the most likely next biosynthetic step in larreatricin (2) metabolism was aromatic ring hydroxylation. In this regard, a protein preparation from L. tridentata was found to convert larreatricin (2), in the presence of ascorbic acid, into two unknown products in an ≈7:1 ratio. A preliminary analysis of the two products (by electron impact-MS and UV) provisionally suggested that both were aromatic hydroxylation products at positions 3′ and 3, i.e., generating compounds 6 and 7 with the former predominating (data not shown).
To unambiguously identify both enzymatic products, 3′ and 3 hydroxylarreatricins (6 and 7) were next synthesized in racemic form (see Supporting Methods and Scheme 2, which is published as supporting information on the PNAS web site). In this context, their structures were established mainly by analysis of their HMBC spectra and NOE patterns (see Supporting Methods and Tables 2 and 3). For (±)-3′-hydroxylarreatricins (6), the catechol ring was established to be connected to C-7′ via the observed cross-peaks from the 7′ proton to carbons 2′ and 6′ at 113.7 and 117.9 ppm (Table 3). Likewise, the phenolic ring was placed at C-7, as evidenced by strong HMBC cross-peaks to carbons 2, 6 at 127.9 ppm. Resonances corresponding to carbons 8 and 9 were also defined via HMBC cross-peak patterns to the 7 proton, and in a similar manner, cross-peaks to the 7′ proton resulted in unambiguous assignments of the 8′ and 9′ positions.
The relative stereochemistry of the furan ring was determined by analysis of the 1D NOESY spectra. Starting with the 7 proton, NOEs were clearly observable with the C-9/C-9′ methyl groups and to the C-2 and C-6 protons. Because of the twisting of the furan ring, however, the 9′ methyl group was in closer proximity to the 7 proton as indicated by the relative intensities of the NOEs to the C-9/C-9′ methyl groups. In addition to the 2′,6′ protons, NOEs resulting from selection of the 7′ proton were observed with the 8 and 8′ protons and to a much lesser extent with the 2 and 6 protons on the phenolic ring. Strong NOE enhancements were additionally observed between the 9 methyl and the 9′ methyl (and vice versa) in agreement with their syn orientation to one another. This, together with the mass spectral fragmentation data, unequivocally established the structures of 6 to be (±)-3′-hydroxylarreatricins. In an analogous manner, the structures of (±)-3-hydroxylarreatricins (7) were also established (Table 2): proof that the phenolic ring was linked to C-7′ was determined through strong cross-peaks from the 7′-proton to the 2′, 6′ carbons at 127.6 ppm, whereas the catechol ring was attached to C-7 as demonstrated by HMBC cross-peaks of the 7-proton to carbons 2 and 6 at 113.7 and 118.1 ppm. The remainder of the structure was verified as previously described for 6. Additionally, in both cases, single frequency decouplings followed by iterative spectral simulation allowed chemical shifts and J couplings to be accurately determined for all coupled protons, with convergence of fit determined by subtraction of simulated from experimental spectra.
With both synthetic 3′-hydroxylarreatricin (6) and 3-hydroxylarreatricin (7) unambiguously identified, attention was next directed to identification of the enzymatic products. Based on HPLC analysis, with UV and electron impact-MS spectra detection and comparison with authentic standards (data not shown), the most abundant enzymatic product was 3′-hydroxylarreatricin (6) and the minor product (7) in a ratio of ≈7:1. Initial attempts to identify the enzymatically generated products by NMR spectroscopy (see Materials and Methods and Table 4) were complicated by only 15 of the 18 carbons being detectable. This complication was caused by both the very low amounts of enzymatic product formed and the signal broadening caused by hydroxyl proton exchange with water in the solvent. However, all resonances of (±)-3′-hydroxylarreatricin (6) were still detectable by analysis of the HMBC data (see Table 3). Nevertheless, these signal detection difficulties were overcome by acquiring the spectrum in dry DMSO-d6 (220 μl) in a microcell NMR tube, this giving an 1D carbon-13 spectrum having all 18 carbons (see Table 4) and the three distinct phenolic protons. Thus, the major enzymatic product was definitively established to be 3′-hydroxylarreatricin 6; in a comparable manner, the minor product was shown to be the 3-hydroxy isomer (7) (data not shown).
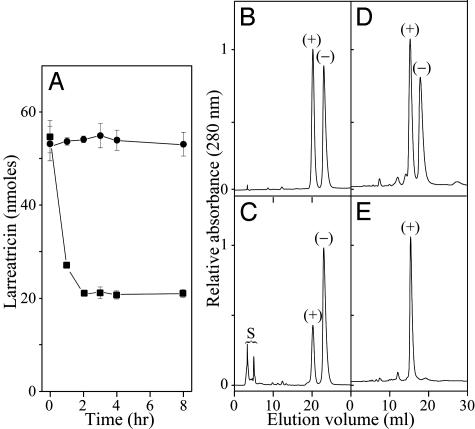
Larreatricin hydroxylase was next purified ≈1,700-fold to apparent homogeneity in ≈7.5% overall yield by DEAE-Sepharose, hydroxylapatite, and Phenyl Sepharose column chromatography and by SDS/PAGE, with the latter suggesting a molecular mass of ≈43 kDa for the native protein. With the purified larreatricin hydroxylase in hand, it was thus next instructive to ascertain whether it catalyzed either regio-specific or enantio-specific aromatic hydroxylation reactions with substrate 2. Thus, (±)-larreatricins (2) were incubated with the purified enzyme preparation, in the presence of ascorbic acid, for time frames ranging from 0 to 8 h (Fig. 2A). After each incubation, the remaining larreatricin (2) substrate and the enzymatically formed 3′-hydroxylarreatricin (6) were individually isolated and subjected to chiral HPLC analysis. [For comparative purposes, Fig. 2 B and D shows the chiral separation of synthetic (+)- and (–)-antipodes of both larreatricin (2) and 3′-hydroxylarreatricin (6), respectively.] After incubation, it was found that only the (+)-larreatricin (2) antipode was depleted during the enzymatic hydroxylation, whereas the (–)-antipode 2 was not (Fig. 2C). In accordance with this observation, the enzymatically formed product 6 was present essentially solely as the (+)-antipode (Fig. 2E). These data thus provided unequivocal proof that the enzymatic transformation was enantio-specific for the (+)-antipode.
Fig. 2.
Enantio-specificity of (+)-larreatricin 3′-hydroxylase. (A) Time course of larreatricin (2) substrate depletion catalyzed by (+)-larreatricin 3′-hydroxylase. ▪, (+)-larreatricin; •, (–)-larreatricin. (B) Chiral separation of synthetic (+)- and (–)-larreatricins (2). (C) Enantiomeric composition of larreatricin (2) after incubation for 4 h with larreatricin 3′-hydroxylase. (D) Chiral separation of synthetic (+)- and (–)-3′-hydroxylarreatricins (6). (E) Enantiomeric composition of enzymatically formed 3′-hydroxylarreatricin (6). S, solvent peak.
Gene Cloning and Characterization. After SDS/PAGE, the purified larreatricin hydroxylase was subjected to proteolytic digestion with separation and analysis of tryptic fragments by microcapillary RP-HPLC and nano-electrospray tandem MS. Two fragments displayed high homology to highly conserved PPO copper binding domains A and B (25, 31–33), including the A and B domains of V. faba PPO (31) [WYLYFYER, DPIFYSHHSNVDR] and the B domain of apricot PPO (32) [DPLFYAHHANVDR]. Degenerate primers were next designed from amino acid sequences, WYLYFYER and DPIFYSHH, with PCR amplification using the L. tridentata cDNA library as template (see Supporting Methods). An internal fragment (464 bp) spanning both domains was thus obtained from which gene-specific reverse and forward primers were designed (see Supporting Methods), and used in PCRs with T3 and T7 primers (universal primers in an Uni-ZAP XR vector), respectively. Although this process resulted in isolation of two PCR fragments (822 bp for the 5′ half and 1,040 bp for the 3′ half) encoding the full-length cDNA, new gene specific primers (forward and reverse, see Supporting Methods) were further used to obtain full-length cDNA (1,796 bp, LtLH). The latter had an ORF of 1,752 bp encoding 584 aa, a stop codon, and 41 bp of a 3′ UTR. The entire deduced amino acid sequence displayed high similarity and identity to apricot (Prunus armeniaca, 60% and 46%) (32), snapdragon (Antirrhinum majus, 59% and 43%) (25), fava bean (Vicia faba, 64% and 51%) (31), and tomato (Lycopersicon esculentum, PPO-E, 56% and 41%) (33) PPOs (Fig. 3); without the N terminus peptide, the similarity/identity to other PPOs (25, 31–33) was 61–67% and 45–67%, respectively. Additionally, the histidine residues at positions 188, 197, 319, 323, and 353 are conserved among the PPOs and are presumed involved in copper binding (34).
Fig. 3.
Alignment of deduced amino acid sequences of cDNA encoding (+)-larreatricin 3′-hydroxylase (LtLH) and those of selected plant PPOs (25, 31–33). Amino acid sequences of peptides derived from purified enzyme are underlined in red. Identical and similar amino acid residues are shown in black and gray, respectively. Copper binding domains A and B are indicated by dark blue and light blue boxes, respectively. Predicted N-terminal and C-terminal processing sites are indicated by arrows.
These data thus confirmed (+)-larreatricin hydroxylase to be an enantio-specific PPO. Moreover, although the gene encoded a protein of ≈66 kDa, the size as estimated by SDS/PAGE was ≈43 kDa, this being a typical feature of PPOs because of posttranslational processing (25, 31–33). That this processing had occurred was confirmed by reanalysis of proteolysis/electrospray fragments originally obtained from purified protein (together with comparison to the deduced amino acid sequence), which revealed posttranslational processing by removal of an 8.7-kDa N-terminal transit peptide (residues 1–79) and an ≈17-kDa COOH-terminal peptide (residues 432–584) (25, 31–33), respectively. Analysis of the transit peptide also revealed that its N-terminal domain is rich in serine residues, followed by a central charged amino acid region, whereas its C-terminal region was hydrophobic, with a putative thylakoid transfer domain (DRRNMLIGLGGLYG) having substantial sequence similarity to other plant PPOs (35). In addition, the transit peptide cleavage site is considered to be between Ala-79 and Ala-80, based on cleavage site motifs proposed by Gavel and von Heijne (36). Indeed, all of these features are common to proteins imported to the thylakoid lumen (37), with further proof of location being determined by immunolocalization studies of PPO by Lax and Vaughn (38) in V. faba. On the other hand, the reason for C-terminal fragment cleavage (between Asn-431 and Asn-432) is unknown, but is speculated as required to obtain biochemically active protein.
PPOs are a widespread class of proteins able to catalyze both one- and two-electron oxidations of various phenols (22–25, 39) to afford corresponding diphenols and quinones, respectively. In terms of physiological roles, many are considered involved in “enzymatic browning” and hence typically thought to have defense functions (40, 41). In general, however, it has been difficult to determine precise physiological functions at the molecular level because of their broad substrate specificities. As summarized earlier, perhaps the most widely claimed function for a PPO involved the in vitro hydroxylation of p-coumaric acid (17) and its glucose/CoA conjugates to afford caffeic acid (18) and its derivatives (23, 42–44). However, this role is suspect for two reasons: first, such PPO-catalyzed hydroxylations can also occur with a broad variety of aromatic substrates (22–25, 39), and second, caffeic acid (18) biosynthesis is now established to be catalyzed by a NADPH-dependent cytochrome P450 (45, 46). Indeed, until the present study, the only other proposed precise physiological roles for PPOs to our knowledge were regio-specific hydroxylation of the achiral 2′,4′,6′,4-tetrahydroxychalcone (19) to give aureusidin (21), and regio-specific dehydrogenation/hydroxylation of 2′,4′,6′,3,4-pentahydroxychalcone (20) to give 21 and 22 in a 6:1 ratio, during yellow flower color development in snapdragon (Antirrhinum majus) (25), as well as regio-specific formation of l-(3,4-dihydroxyphenyl)-alanine (Dopa, 24) from tyrosine (23) and of cyclo-Dopa (26) via Dopaquinone (25), the precursors of the Portulaca grandiflora betalains (47) (Scheme 1). Note, however, that the snapdragon PPO transformations were also demonstrated to be catalyzed by a Neurospora crassa tyrosinase, a nonspecific binuclear Cu-containing PPO, suggesting that such conversions lack strict enzyme-substrate specificity (25). Additionally, the tyrosine-hydroxylating enzyme present in P. grandiflora root tissue was able to use both d- and l-tyrosine, with the latter apparently only very slightly preferred, i.e., there was no strict specificity for either form. Accordingly, the study of creosote bush lignan biosynthesis now offers an additional dimension to the role of PPOs by demonstration of this enantio-specific role in aromatic ring hydroxylations. This enantio-specificity is also of interest, given that one of the major lignan end-products in this species is actually the optically inactive NDGA (10), whose formation involves both regio-selective (8–8′) coupling, and the downstream PPO-catalyzed enantio-specific conversions. These findings can thus be anticipated to give additional insight and direction to the study of plant PPOs in general and should provide further impetus to establish their actual physiological roles in vivo.
Supplementary Material
Acknowledgments
We thank Professor Daneel Ferreira for high-resolution mass spectroscopic analyses. We gratefully acknowledge the National Science Foundation (Grant MCB9976684), the U.S. Department of Energy (Grant DE FG03-97ER20259), McIntire Stennis, the Lewis B. and Dorothy Cullman and G. Thomas Hargrove Center for Land Plant Adaptation Studies, and the G. Thomas and Anita Hargrove Center for Plant Genomic Research for generous support of this study.
Abbreviations: PPO, polyphenol oxidase; NDGA, nordihydroguaiaretic acid; NOE, nuclear Overhauser effect; HMBC, heteronuclear multibond connectivity.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AY370019).
References
- 1.Train, P., Henrichs, J. R. & Archer, W. A. (1941) Medicinal Uses of Plants by Indian Tribes of Nevada (U.S. Department of Agriculture, Washington, DC).
- 2.Waller, C. W. & Gisvold, O. (1945) J. Am. Pharm. Assoc. 34, 78–81. [Google Scholar]
- 3.Timmermann, B. N. (1977) in Creosote Bush: Biology and Chemistry of Larrea in New World Deserts, eds. Mabry, T. J., Hunziker, J. H. & DiFeo, D. R., Jr. (Dowden, Hutchinson & Ross, Stroudsburg, PA), pp. 252–276.
- 4.Lundberg, W. O., Halvorson, H. O. & Burr, G. O. (1944) Oil Soap 21, 33–35. [Google Scholar]
- 5.Oliveto, E. P. (1972) Chem. Industry 677–679.
- 6.Belmares, H., Barrera, A., Castillo, E., Ramos, L. F., Hernandez, F. & Hernandez, V. (1979) Ind. Eng. Chem. Prod. Res. Dev. 18, 220–226. [Google Scholar]
- 7.Luo, J., Chuang, T., Cheung, J., Quan, J., Tsai, J., Sullivan, C., Hector, R. F., Reed, M. J., Meszaros, K., King, S. R., et al. (1998) Eur. J. Pharmacol. 346, 77–79. [DOI] [PubMed] [Google Scholar]
- 8.Lintner, K. (2000) International Patent WO 2000028959.
- 9.Elakovich, S. D. & Stevens, K. L. (1985) J. Chem. Ecol. 11, 27–33. [DOI] [PubMed] [Google Scholar]
- 10.McDonald, R. W., Bunjobpon, W., Liu, T., Fessler, S., Pardo, O. E., Freer, I. K., Glaser, M., Seckl, M. J. & Robins, D. J. (2001) Anticancer Drug Des. 16, 261–270. [PubMed] [Google Scholar]
- 11.Gnabre, J. N., Brady, J. N., Clanton, D. J., Ito, Y., Dittmer, J., Bates, R. B. & Huang, R. C. C. (1995) Proc. Natl. Acad. Sci. USA 92, 11239–11243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Craigo, J., Callahan, M., Huang, R. C. C. & DeLucia, A. L. (2000) Antiviral Res. 47, 19–28. [DOI] [PubMed] [Google Scholar]
- 13.Gnabre, J. N., Ito, Y., Ma, Y. & Huang, R. C. (1996) J. Chromatogr. A 719, 353–364. [DOI] [PubMed] [Google Scholar]
- 14.Konno, C., Xue, H.-Z., Lu, Z.-Z., Ma, B.-X., Erdelmeier, C. A. J., Che, C.-T., Cordell, G. A., Soejarto, D. D., Waller, D. P. & Fong, H. H. S. (1989) J. Nat. Prod. 52, 1113–1117. [DOI] [PubMed] [Google Scholar]
- 15.Konno, C., Lu, Z.-Z., Xue, H.-Z., Erdelmeier, C. A. J., Meksuriyen, D., Che, C.-T., Cordell, G. A., Soejarto, D. D., Waller, D. P. & Fong, H. H. S. (1990) J. Nat. Prod. 53, 396–406. [DOI] [PubMed] [Google Scholar]
- 16.Moinuddin, S. G. A., Hishiyama, S., Cho, M.-H., Davin, L. B. & Lewis, N. G. (2003) Org. Biomol. Chem. 1, 2307–2313. [DOI] [PubMed] [Google Scholar]
- 17.Croteau, R., Kutchan, T. M. & Lewis, N. G. (2000) in Biochemistry and Molecular Biology of Plants, eds. Buchanan, B. B., Gruissem, W. & Jones, R. L. (Am. Soc. Plant Physiol., Rockville, MD), pp. 1250–1318.
- 18.Lewis, N. G. & Davin, L. B. (1999) in Comprehensive Natural Products Chemistry, eds. Barton, D. H. R., Nakanishi, K. & Meth-Cohn, O. (Elsevier, London), Vol. 1, pp. 639–712. [Google Scholar]
- 19.Davin, L. B., Wang, H.-B., Crowell, A. L., Bedgar, D. L., Martin, D. M., Sarkanen, S. & Lewis, N. G. (1997) Science 275, 362–366. [DOI] [PubMed] [Google Scholar]
- 20.Gang, D. R., Costa, M. A., Fujita, M., Dinkova-Kostova, A. T., Wang, H.-B., Burlat, V., Martin, W., Sarkanen, S., Davin, L. B. & Lewis, N. G. (1999) Chem. Biol. 6, 143–151. [DOI] [PubMed] [Google Scholar]
- 21.Halls, S. C. & Lewis, N. G. (2002) Biochemistry 41, 9455–9461. [DOI] [PubMed] [Google Scholar]
- 22.Butt, V. S. (1977) in Recent Advances in Phytochemistry, eds. Swain, T., Harborne, J. B. & van Sumere, C. F. (Plenum, New York), Vol. 12, pp. 433–456. [Google Scholar]
- 23.Kojima, M. & Takeuchi, W. (1989) J. Biochem. 105, 265–270. [DOI] [PubMed] [Google Scholar]
- 24.Billaud, C., Lecornu, D. & Nicolas, J. (1996) J. Agric. Food Chem. 44, 1668–1675. [Google Scholar]
- 25.Nakayama, T., Yonekura-Sakakibara, K., Sato, T., Kikuchi, S., Fukui, Y., Fukuchi-Mizutani, M., Ueda, T., Nakao, M., Tanaka, Y., Kusumi, T. & Nishino, T. (2000) Science 290, 1163–1166. [DOI] [PubMed] [Google Scholar]
- 26.Halls, S. C. & Lewis, N. G. (2003) Tetrahedron Assym. 14, 649–658. [Google Scholar]
- 27.Bradford, M. M. (1976) Anal. Biochem. 72, 248–254. [DOI] [PubMed] [Google Scholar]
- 28.Eng, J. K., McCormack, A. L. & Yates, J. R., III. (1994) J. Am. Soc. Mass Spectrom. 5, 976–989. [DOI] [PubMed] [Google Scholar]
- 29.Chittum, H. S., Lane, W. S., Carlson, B. A., Roller, P. P., Lung, F. D., Lee, B. J. & Hatfield, D. L. (1998) Biochemistry 37, 10866–10870. [DOI] [PubMed] [Google Scholar]
- 30.Taniguchi, T., Garcia-Higuera, I., Xu, B., Andreassen, P. R., Gregory, R. C., Kim, S. T., Lane, W. S., Kastan, M. B. & D'Andrea, A. D. (2002) Cell 109, 459–472. [DOI] [PubMed] [Google Scholar]
- 31.Cary, J. W., Lax, A. R. & Flurkey, W. H. (1992) Plant Mol. Biol. 20, 245–253. [DOI] [PubMed] [Google Scholar]
- 32.Chevalier, T., de Rigal, D., Mbéguié-A-Mbéguié, D., Gauillard, F., Richard-Forget, F. & Fils-Lycaon, B. R. (1999) Plant Physiol. 119, 1261–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Newman, S. M., Eannetta, N. T., Yu, H., Prince, J. P., de Vincente, M. C., Tanksley, S. D. & Steffens, J. C. (1993) Plant Mol. Biol. 21, 1035–1051. [DOI] [PubMed] [Google Scholar]
- 34.Klabunde, T., Eicken, C., Sacchettini, J. C. & Krebs, B. (1998) Nat. Struct. Biol. 5, 1084–1090. [DOI] [PubMed] [Google Scholar]
- 35.Joy, R. W., IV, Sugiyama, M., Fukuda, H. & Komamine, A. (1995) Plant Physiol. 107, 1083–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gavel, Y. & von Heijne, G. (1990) FEBS Lett. 261, 455–458. [DOI] [PubMed] [Google Scholar]
- 37.Smeekens, S., Bauerle, C., Hageman, J., Keegstra, K. & Weisbeek, P. (1986) Cell 46, 365–375. [DOI] [PubMed] [Google Scholar]
- 38.Lax, A. R. & Vaughn, K. C. (1991) Plant Physiol. 96, 26–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rompel, A., Fischer, H., Meiwes, D., Büldt-Karentzopoulos, K., Magrini, A., Eicken, C., Gerdemann, C. & Krebs, B. (1999) FEBS Lett. 445, 103–110. [DOI] [PubMed] [Google Scholar]
- 40.Thipyapong, P., Hunt, M. D. & Steffens, J. C. (1995) Phytochemistry 40, 673–676. [Google Scholar]
- 41.Constabel, C. P., Yip, L., Patton, J. J. & Christopher, M. E. (2000) Plant Physiol. 124, 285–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vaughan, P. F. T. & Butt, V. S. (1969) Biochem. J. 113, 109–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tanaka, M. & Kojima, M. (1991) Arch. Biochem. Biophys. 284, 151–157. [DOI] [PubMed] [Google Scholar]
- 44.Wang, Z.-X., Li, S.-M., Löscher, R. & Heide, L. (1997) Arch. Biochem. Biophys. 347, 249–255. [DOI] [PubMed] [Google Scholar]
- 45.Schoch, G., Goepfert, S., Morant, M., Hehn, A., Meyer, D., Ullmann, P. & Werck-Reichhart, D. (2001) J. Biol. Chem. 276, 36566–36574. [DOI] [PubMed] [Google Scholar]
- 46.Franke, R., Humphreys, J. M., Hemm, M. R., Denault, J. W., Ruegger, M. O., Cusumano, J. C. & Chapple, C. (2002) Plant J. 30, 33–45. [DOI] [PubMed] [Google Scholar]
- 47.Strack, D. & Schliemann, W. (2001) Angew. Chem. Int. Ed. 40, 3791–3794. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.