Abstract
Sterols mediate feedback inhibition of the sterol regulatory element-binding protein (SREBP) pathway by preventing movement of the SREBP cleavage-activating protein (SCAP)/SREBP complex from endoplasmic reticulum (ER) to Golgi, where proteolytic cleavage of SREBPs releases the transcription factor domain that activates genes for lipid biosynthesis. Our laboratory previously used a trypsin cleavage assay to show that the conformation of SCAP is altered in vitro by addition of cholesterol to ER membranes. More recently, Insig-1 and Insig-2 were identified as ER resident proteins that bind the SCAP/SREBP complex and promote its ER retention when cells are treated with sterols. Here, we use the trypsin assay to show that Insig proteins reduce the concentration of cholesterol needed in vitro to produce the conformational change in SCAP. Insig-1 and Insig-2 also enhance the conformational change in SCAP that occurs upon addition of certain cationic amphiphiles, such as chlorpromazine, trifluoperazine, and imipramine, which mimic the effect of cholesterol. The effects of cationic amphiphiles raise the possibility that SCAP may monitor specifically the composition of the cytoplasmic leaflet of the ER membrane.
Sterol regulatory element-binding proteins (SREBPs) are a family of transcription factors that control lipid homeostasis in animal cells (1). During synthesis, SREBPs are inserted into the membranes of the endoplasmic reticulum (ER) in a helical hairpin fashion with both the NH2- and COOH-terminal domains facing the cytosolic side (2). The NH2-terminal domain comprises a basic helix–loop–helix leucine-zipper transcription factor, but it remains inactive while tethered to the membrane. Activation of SREBPs requires a second polytopic membrane protein called SREBP cleavage-activating protein (SCAP). SCAP associates with SREBPs immediately after their synthesis in the ER and escorts SREBPs to the Golgi where SREBPs are sequentially cleaved by two proteases. Proteolysis releases the transcription factor domain of SREBP so that it may travel to the nucleus and activate the entire program of genes required for cholesterol and fatty acid synthesis.
In conjunction with its role as an escort protein, SCAP is required for feedback inhibition of the SREBP pathway by sterols (3). When sterols are present at high concentrations in a cell, the SCAP/SREBP complex is retained in the ER. When a cell is depleted of sterols, SCAP escorts SREBP to the Golgi for proteolytic processing. SCAP contains eight transmembrane segments (TMs). TMs 2–6, which comprise the sterol-sensing domain, appear crucial for sterol regulation: three different missense mutations in this domain render SCAP insensitive to sterols (4–6).
An important unanswered question is whether SCAP directly senses the sterol content of the ER membrane or whether the signal is transmitted by more indirect means. This laboratory previously reported a trypsin cleavage assay designed to test the effect of sterols on SCAP conformation in vitro (7). In this assay, SCAP-containing membrane vesicles are isolated from cells and treated briefly with trypsin, which digests the cytoplasmic side of the vesicles while leaving the TMs and luminal domains intact. To monitor changes in SCAP conformation, we use a mAb that specifically recognizes the fourth ER luminal loop of SCAP, between TMs 7 and 8. Under control conditions, trypsin cleaves SCAP on the cytoplasmic sides of TM 7 (arginine residue 496) and TM 8 (arginines 747–750), generating a protected 250-aa fragment that can be detected by immunoblot using the anti-SCAP antibody. If membranes are incubated in vitro with cholesterol in complex with methyl-β-cyclodextrin (MCD) before trypsin treatment, two arginines closer to the membrane (residues 503 and 505) become accessible to trypsin, and thus a smaller 241- to 243-aa fragment is detected. This shift in arginine accessibility indicates that cholesterol alters SCAP conformation, and this alteration may lead to SCAP retention in the ER.
More recently, two proteins, Insig-1 and Insig-2, were shown to cooperate with sterols to inhibit exit of the SCAP/SREBP complex from the ER (8, 9). Insig-1 and Insig-2 are closely related polytopic membrane proteins that remain in the ER and interact with TMs 1–6 of SCAP in a sterol-dependent manner to form a ternary SCAP/SREBP/Insig complex. In the absence of sterols, SCAP does not interact with Insig proteins. As a result, the SCAP/SREBP complex exits the ER, and SREBP is delivered to the Golgi. In the presence of sterols, SCAP binds to Insigs, and this binding is essential for ER retention of the SCAP/SREBP complex. When SCAP bears one of the three sterol-sensing domain mutations, its binding to Insig is disrupted, and ER retention does not occur (8, 9). Defective regulation also occurs when SCAP is overexpressed to such a high level that Insig becomes saturated (8). Thus, a high ratio of SCAP to Insig diminishes sterol sensitivity of SREBP processing. Conversely, as Insig levels rise, SREBP processing is inhibited by lower concentrations of sterols. These data suggest that the SCAP/Insig complex has a higher affinity for cholesterol as compared with SCAP alone.
In the current experiments, we use the trypsin cleavage assay to test the effect of Insig proteins on the sensitivity of SCAP to the addition of cholesterol in vitro. We also test the hypothesis that the conformation of SCAP might also be altered by changes in the lipid environment of the ER membrane. For this purpose, we examine the effect of cationic amphiphiles that are known to perturb membrane structure (10–12).
Materials and Methods
Materials used in this study and certain methods (construction of expression plasmids and immunoblot analysis) may be found in Supporting Text, which is published as supporting information on the PNAS web site, www.pnas.org.
Tissue Culture Media. Medium A is a 1:1 mixture of Ham's F-12 medium and DMEM containing 100 units/ml penicillin and 100 μg/ml streptomycin sulfate. Medium B is medium A supplemented with 5% (vol/vol) FCS, 5 μg/ml cholesterol, 1 mM sodium mevalonate, and 20 μM sodium oleate. Medium C is medium A supplemented with 5% newborn calf lipoprotein-deficient serum, 50 μM sodium compactin, and 50 μM sodium mevalonate.
Culture, Transfection, and Fractionation of SRD-13A Cells. SRD-13A cells are a SCAP null mutant clone derived from γ-irradiated Chinese hamster ovary-7 cells (13). Cell monolayers were maintained in medium B at 37°C in 8–9% CO2. On day 0, cells were set up for experiments in medium B at 7.5 × 105 cells per 100-mm dish. On day 2, the cells were transfected with plasmids by using FuGENE 6 reagent as described (6). The total amount of DNA in each transfection was adjusted to 5–7 μg per dish by addition of pcDNA3 mock vector. After transfection, cells were incubated at 37°C for 8–12 h in medium A supplemented with 5% FCS and then switched to medium C. After incubation for 16 h, the cells were harvested, and a 20,000 × g membrane fraction was prepared as described (7).
Trypsin Cleavage Assay of SCAP. Our protocol was adapted from Brown et al. (7) with modifications designed to maximize the effect of Insig on SCAP's sensitivity to cholesterol. Aliquots (100 μg of protein) of the 20,000 × g membrane fraction from SRD-13 cells were resuspended in buffer A (10 mM Hepes·KOH, pH 7.4/10 mM KCl/1.5 mM MgCl2/5 mM sodium EDTA/5 mM sodium EGTA/250 mM sucrose) in the absence or presence of the sterol/MCD complex or drug to give a final volume of 500 μl. The mixture was incubated at room temperature for 20 min, then centrifuged at 20,000 × g for 10 min at 4°C. Buffer was aspirated from the resulting membrane pellet, and the membranes were resuspended in 136 μl of buffer A. To each suspension was added 1.6 μg of trypsin (in 4 μl), and the samples were incubated at 30°C for 30 min. Trypsin digestion was stopped by addition of 80 μg of soybean trypsin inhibitor (in 4 μl), after which the membrane suspension was subjected to PNGase F treatment (37°C for 4–12 h) and acetone precipitation as described (7). Air-dried precipitated samples were dissolved in 100 μl of SDS loading buffer and then subjected to SDS/PAGE and immunoblot analysis.
Results
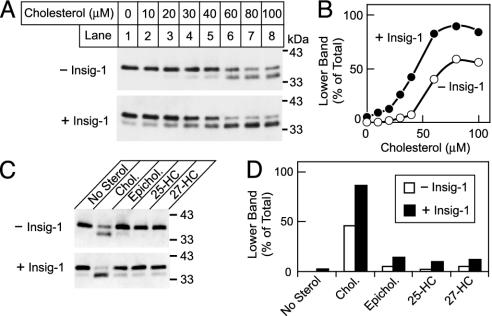
To test the hypothesis that Insig-1 might increase the sensitivity of SCAP to cholesterol, we controlled the expression of SCAP by using a SCAP-deficient Chinese hamster ovary cell line (SRD-13A) (13). The cells were transfected with cDNAs encoding SCAP and SREBP-2 as well as various amounts of cDNA encoding Insig-1. The cells were then incubated in sterol-depleting conditions, and a membrane fraction containing ER was isolated. The membranes were incubated in vitro with cholesterol in complex with MCD. SCAP conformation was analyzed by trypsin cleavage followed by SDS/PAGE and immunoblot analysis with mAb IgG-9D5. In the absence of cholesterol, trypsin cleaved SCAP to a protected fragment with an apparent molecular mass of 37 kDa (Fig. 1A Upper, lane 1). If cholesterol was added to the membranes in a sufficiently high concentration before trypsin treatment, a smaller protected fragment was seen (35 kDa; Fig. 1 A Upper, lane 8). The smaller protected fragment reflects the conformational change in SCAP that exposes arginine-503/arginine-505 (R503/R505) to trypsin cleavage (7). The apparent molecular masses of the upper and lower bands in Fig. 1 A were higher than those observed in our earlier studies (37 vs. 27 kDa and 35 vs. 26 kDa), owing to the current use of a Tris-tricine gel buffer system vs. the earlier use of Tris-glycine (7).
Fig. 1.
Insig-1 enhances SCAP's response to in vitro treatment with cholesterol. (A) SRD-13A cells were transfected with 2 μg of pCMV (cytomegalovirus)-SCAP and 2 μg of pTK-HSV-SREBP-2 in the absence or presence of 0.3 μg of pCMV-Insig-1-Myc as indicated. After the cells were harvested, aliquots of the 20,000 × g membrane suspension (100 μg) were incubated for 20 min at room temperature with the indicated concentration of cholesterol/MCD complex. At the end of the incubation, the membranes were treated sequentially with trypsin (30°C for the 30 min) and PNGase F and were then subjected to SDS/PAGE and immunoblot analysis with anti-SCAP IgG-9D5. (B) Quantification of Insig-1's effect on SCAP's response to cholesterol. The relative intensity of the upper and lower bands in A was quantified by densitometry. (C) Insig-1 does not enhance SCAP's sensitivity to epicholesterol or oxysterols. SRD-13A cells were transfected with 2 μg of pCMV-SCAP and 2 μg of pTK-HSV-SREBP-2 in the absence or presence of 0.3 μg of pCMV-Insig-1-Myc as indicated. Aliquots of the 20,000 × g membrane suspension were incubated with 80 μM of the indicated sterol/MCD complex. After sterol treatment, SCAP's conformation was analyzed as described in A.(D) Relative intensity of the upper and lower bands in C was quantitated by densitometry. Chol. cholesterol; epichol., epicholesterol; 25-HC, 25-hydroxycholesterol; 27-HC, 27-hydroxycholesterol.
Fig. 1 A also shows that cotransfection of cells with Insig-1 enhanced the sensitivity of SCAP to the addition of cholesterol in vitro. The relative amounts of upper (37 kDa) and lower (35 kDa) bands of trypsin-treated SCAP were measured densitometrically, and the amount of the lower band was expressed as a percent of the total. As cholesterol was added to the membrane, the percent of the lower band increased in a sigmoidal fashion. At a maximum, ≈50% of SCAP was susceptible to cleavage at R503/R505 (Fig. 1B). When Insig-1 was overexpressed, the cholesterol curve shifted to the left, and ≈100% of SCAP molecules could be cleaved at R503/R505. Results similar to those in Fig. 1 A were seen in 10 other experiments.
In intact mammalian cells, oxysterols are potent suppressors of SREBP processing, and Insig proteins increase cellular sensitivity to oxysterols (8). However, oxysterols do not alter SCAP conformation when delivered to the ER membrane in vitro (7). We asked whether Insig-1 might allow oxysterols in vitro to alter the conformation of SCAP. We tested oxysterols at 80 μM, which is a saturating concentration for cholesterol. Fig. 1 C and D shows that SCAP conformation was not affected by 80 μM 25-hydroxycholesterol or 27-hydroxycholesterol, even when Insig-1 was coexpressed. SCAP conformation was also insensitive to epicholesterol, the 3α stereoisomer of cholesterol.
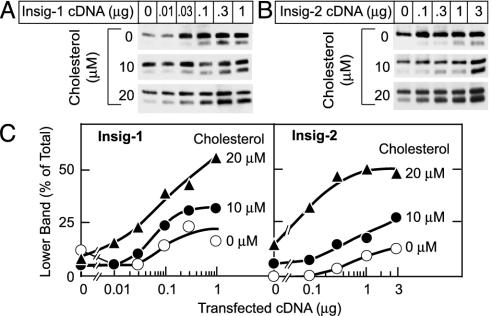
In intact cells, Insig-2 resembles Insig-1 in its capacity to interact with SCAP and sensitize SREBP processing to sterols (9). Fig. 2 shows that the response of SCAP to submaximal doses of cholesterol in vitro increased with increasing amounts of transfected Insig-2 cDNA (B and D) in the same manner as with Insig-1 cDNA (A and C). Thus, both Insig isoforms enhanced the sensitivity of SCAP to cholesterol in vitro, consistent with their capacity to increase the sterol sensitivity of SREBP processing.
Fig. 2.
Both Insig-1 and Insig-2 enhance SCAP's response to cholesterol. SRD-13A cells were transfected with 2 μg of pCMV-SCAP, 2 μg of pTK-HSV-SREBP-2, and the indicated amounts of pCMV-Insig-1-Myc (A) or pCMV-Insig-2-Myc (B). Aliquots of 20,000 × g membrane suspension were incubated for 20 min at room temperature with the indicated concentration of cholesterol/MCD complex. After sterol treatment, SCAP's conformation was analyzed as described in Fig. 1. (C) Relative intensity of the upper and lower bands in A and B was quantified by densitometry.
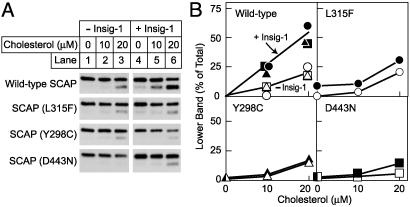
The binding of SCAP to Insig proteins is abolished by any one of three missense mutations (Y298C, L315F, or D443N) in SCAP that alter residues that are predicted to lie near the cytoplasmic boundaries of TMs 2, 3, and 6, respectively (4–6). Each of these three mutations was originally identified by its capacity to render SREBP processing resistant to sterol suppression in intact cells. Fig. 3A shows that the Y298C, L315F, and D443N mutations each rendered SCAP insensitive to the Insig-mediated enhancement of cholesterol sensitivity. This is most apparent at 20 μM cholesterol (Fig. 3A, lane 6), and the conclusion was supported by densitometry (Fig. 3B). This result suggests that Insig-1 must bind SCAP to enhance cholesterol sensitivity and that the sterol-sensing mutations in SCAP interfere with this binding.
Fig. 3.
Sterol-resistant mutant forms of SCAP show diminished Insig-1-dependent response to cholesterol. (A) SRD-13A cells were transfected with 2μg of the indicated SCAP construct plus 2 μg of pTK-HSV-SREBP-2 in the absence or presence of 0.1 μg of pCMV-Insig-1-Myc as indicated. Aliquots of the 20,000 × g membrane suspension were incubated for 20 min at room temperature with the indicated concentration of the cholesterol/MCD complex. After sterol treatment, SCAP's conformation was analyzed as described in Fig. 1. (B) Relative intensity of the top and bottom bands in A was quantified by densitometry. Data are from three independent experiments comparing wild-type SCAP to a SCAP mutant: wild type vs. L315F (circles), wild type vs. Y298C (triangles), and wild type vs. D443N (squares). The wild-type SCAP immunoblots in A are from the experiment comparing wild type to SCAP (L315F).
The effect of cholesterol on SCAP conformation might reflect direct binding between cholesterol and SCAP. Alternatively, cholesterol might affect SCAP by altering ER lipid composition and physical properties. One piece of evidence suggests that SCAP is sensitive to lipid composition. In Drosophila cells, SREBP processing requires SCAP (14), but it is inhibited by phosphatidylethanolamine (PE), not sterols (15). Under certain conditions, PE and cholesterol can have similar effects on membrane physical properties (reviewed in ref. 15).
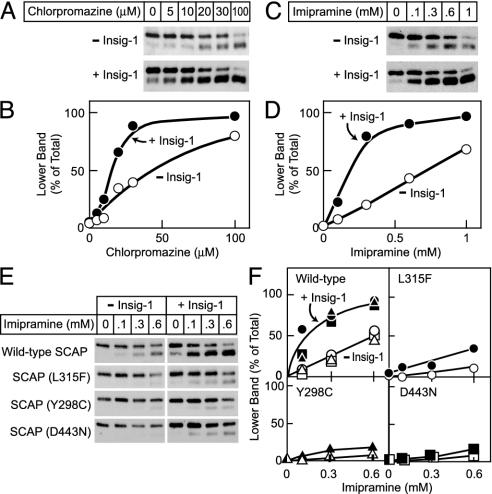
To test the hypothesis that the conformation of mammalian SCAP might change in response to membrane properties, we tested the effects of chlorpromazine and imipramine, two prototypical cationic amphiphiles that are known to perturb membrane structure (10, 16–18). We asked whether these compounds might alter SCAP conformation in the in vitro trypsin digestion assay and whether the response to these drugs might be enhanced by Insig-1 coexpression. Fig. 4 shows that both chlorpromazine (A and B) and imipramine (C and D) altered SCAP conformation as indicated by an increased generation of the lower trypsin-protected band. The dose–response curves (Fig. 4 B and D) demonstrate that the major difference between these two compounds lies in their effective concentrations. Both compounds were able to convert the majority of SCAP to the lower band even in the absence of Insig-1. Chlorpromazine achieved this effect at 100 μM, whereas imipramine required a 10-fold higher concentration (note the difference in scales on the horizontal axis of Fig. 4 B and D). Insig-1 markedly enhanced the response to chlorpromazine or imipramine when they were tested at submaximal concentrations.
Fig. 4.
Insig-1 sensitizes wild-type but not sterol-resistant SCAP to chlorpromazine and imipramine. SRD-13A cells were transfected with 2 μg of wild type or mutant pCMV-SCAP and 2 μg of pTK-HSV-SREBP-2 in the absence or presence of 0.3 μg of pCMV-Insig-1-Myc as indicated. Aliquots of the 20,000 × g membrane suspension were incubated for 20 min at room temperature with the indicated concentration of chlorpromazine or imipramine. After treatment with drug, SCAP's conformation was analyzed as described in Fig. 1. (A) Effect of varying concentrations of chlorpromazine on wild-type SCAP. (B) Quantification of data in A by densitometry. (C) Effect of varying concentrations of imipramine on wild-type SCAP. (D) Quantification of data in C by densitometry. (E) Effect of varying concentrations of imipramine on sterol-resistant mutant forms of SCAP. (F) Relative intensity of the top and bottom bands in E was quantified by densitometry. Data are from three independent experiments comparing wild-type SCAP to a SCAP mutant: wild type vs. L315F (circles), wild type vs. Y298C (triangles), and wild type vs. D443N (squares). The wild-type SCAP immunoblots in E are from the experiment comparing wild type to SCAP (L315F).
As discussed above for Fig. 3, Insig-1 had a diminished effect on the response of SCAP to cholesterol when SCAP contained mutations Y298C, L315F, or D443N. Fig. 4 E and F shows that these mutations also diminished the response of SCAP to imipramine and blocked the ability of Insig-1 to increase this response.
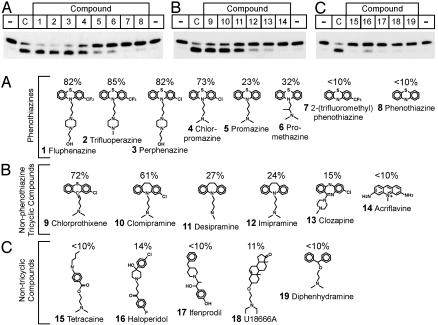
To determine the important structural features of chlorpromazine and imipramine, we tested 17 additional compounds at concentrations of 100 μM in the presence of Insig-1. Fig. 5A shows eight phenothiazine compounds, of which chlorpromazine is the prototype. Chlorpromazine (compound 4) consists of a tricyclic phenothiazine group, an electronegative chlorine atom at position 2 of the phenothiazine group, and an aliphatic side chain at position 10 that terminates in a tertiary amine group. At 100 μM, chlorpromazine generated the same amount of lower band as did a saturating dose of cholesterol (80 μM). A similar effect was observed with trifluoperazine (2), fluphenazine (1), and perphenazine (3). All three of these phenothiazines have electronegative side groups (-C1 or –CF3) and an amine-containing piperazine side chain. The relative effect on SCAP conformation was markedly reduced by an absence of the electronegative group, as illustrated by promazine (5) and promethazine (7), and was abolished by absence of the amine-containing side chain, as illustrated by 2-trifluoromethylphenothiazine (7) and phenothiazine (8).
Fig. 5.
Structure-function analysis of compounds that alter SCAP conformation. SRD-13A cells were transfected with 2 μg of pCMV-SCAP, 2 μg of pTK-HSV-SREBP-2, and 0.3 μg of pCMV-Insig-1. Aliquots of the 20,000 × g membrane suspension were incubated for 20 min at room temperature with 80 μM of cholesterol/MCD complex (C) or 100 μM of the indicated compound (numbered 1–19). Stock solutions of drugs were freshly prepared in water (1, 2, 4–6, 10–12, 14, 15, and 17–19) or ethanol (3, 7–9, 13, and 16). After treatment with drug, SCAP's conformation was analyzed by trypsin digestion and immunoblot analysis as described in Fig. 1. The first and last lanes in each gel represent membranes treated with water or ethanol, respectively, in the absence of drug. The percent lower band generated by each compound is shown above the compound's chemical structure and represents the average of at least three experiments.
Fig. 5B shows six nonphenothiazine tricyclic compounds that were tested. This group includes imipramine (12), which consists of a tricyclic dibenzazepine ring and an aliphatic side chain identical to that of chlorpromazine. At 100 μM, imipramine has a small effect on SCAP conformation (24% lower band). Consistent with our studies of the phenothiazines, addition of an electronegative chlorine atom to the dibenzazepine ring, yielding clomipramine (10), significantly increased the effect on SCAP conformation. A significant effect was also observed with chlorprothixene (9), a compound identical to chlorpromazine but with a thioxanthene group in place of the phenothiazine group. Desipramine (11), similar to imipramine but with a secondary amine in place of the tertiary amine, had a greater effect than imipramine. Two additional tricyclic compounds, clozapine (13) and acriflavine (14), had minimal effect at 100 μM (Fig. 5B), as did five cationic amphiphiles compounds that lack a tricyclic ring structure, including tetracaine (15), haloperidol (16), ifenprodil (17), U18666A (18), and diphenhydramine (19) (Fig. 5C).
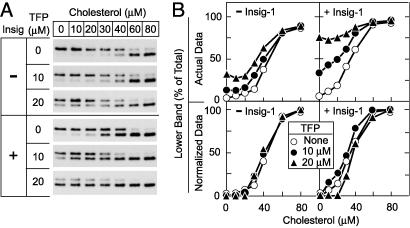
To determine whether the effects of cholesterol and cationic amphiphiles are additive or synergistic, we assessed the effect of trifluoperazine on SCAP's response to cholesterol (Fig. 6). We chose trifluoperazine because it had the greatest effect of the compounds examined in Fig. 5 and was slightly more potent than chlorpromazine in more detailed dose–response curves (data not shown). In the absence of cholesterol, low concentrations of trifluoperazine (10 or 20 μM) slightly increased the amount of lower band in the absence of Insig-1 and had a greater effect in the presence of Insig-1. In the absence of trifluoperazine, addition of a submaximal concentration of cholesterol (30 μM) increased the amount of lower band, most prominently in the presence of Insig-1. Further addition of trifluoperazine produced an additive effect. The additivity of cholesterol and trifluoperazine raises the possibility that both compounds act through the same mechanism.
Fig. 6.
Additive effects of Insig-1 and trifluoperazine (TFP) on SCAP's response to cholesterol. (A) SRD-13A cells were transfected with 2 μg of pCMV-SCAP and 2 μg of pTK-HSV-SREBP-2, in the absence or presence of 0.3 μg of pCMV-Insig-1-Myc as indicated. Aliquots of the 20,000 × g membrane suspension were incubated for 20 min at room temperature with the indicated concentration of cholesterol/MCD complex alone (○) or in the presence of TFP at 10 μM(•) or 20 μM(▴). After treatment, SCAP's conformation was analyzed as described in Fig. 1. (B) Relative intensity of the top and bottom bands in A was quantified by densitometry. The actual data in A and B were normalized in Lower as follows: for each cholesterol curve, the value for % lower band generated in the absence of cholesterol and in the presence of 0, 10, or 20 μM TFP was subtracted from all other points on the curve, after which the subtracted values for each curve were normalized to the maximal value at 80μM cholesterol, which was set at 100%.
Discussion
Insig proteins were recently identified as important regulatory participants in the SREBP pathway (8, 9). Sterols and Insig proteins function together to cause retention of the SCAP/SREBP complex in the ER. If intact cells are treated with sterols or if Insig levels are high, the SCAP/SREBP complex remains in the ER, bound to Insig. Conversely, when cells are depleted of sterols or when Insig levels are low, the SCAP/SREBP complex is released to the Golgi. There, SREBP is proteolytically cleaved to activate the genes responsible for cholesterol and fatty acid synthesis.
The mechanism by which sterols and Insig proteins cause ER retention of the SCAP/SREBP complex is beginning to be revealed. We previously reported that treatment of intact cells with 25-hydroxycholesterol or treatment of ER membranes in vitro with cholesterol causes a conformational change in SCAP, exposing R503/R505 to cleavage by trypsin (7). This sterol-dependent conformational change in SCAP does not appear to have an absolute requirement for Insig, based on the finding that mammalian SCAP expressed in Sf9 insect cells shows a similar cholesterol-induced conformational change in vitro (7). Insect genomes do not encode Insigs as revealed by computer searches of the sequences of Drosophila melanogaster and Drosophila pseudoobscura.
In the current article, we used the in vitro trypsin cleavage assay to test how Insig proteins affect the changes in SCAP conformation in response to sterols. Our data provide three insights into the mechanism by which sterols and Insig mediate ER retention of SCAP. First, we found that coexpression of Insig with SCAP significantly lowers the amount of cholesterol required to change the conformation of SCAP. These data are consistent with previous studies of intact cells in which Insig expression decreased the concentration of 25-hydroxycholesterol or low-density lipoprotein-derived cholesterol required to cause ER retention of the SCAP/SREBP complex (8, 9). The effect of Insig in vitro was diminished by mutations in the sterol-sensing domain of SCAP that reduce SCAP binding to Insig. Taken together, the data suggest a model in which sterols alter the conformation of SCAP, thereby promoting its association with Insig. The SCAP/Insig complex may retain cholesterol more tightly than free SCAP. This mechanism would explain the observation that Insig shifts the sterol response curve to the left, whether we measure SREBP processing in vivo or trypsin sensitivity of SCAP in vitro.
The second conclusion relates to the question of which sterol interacts with SCAP in the ER membrane of intact cells. When added to the extracellular medium of intact cells, oxysterols, such as 25-hydroxycholesterol or 27-hydroxycholesterol, are significantly more potent than cholesterol in causing ER retention of the SCAP/SREBP complex (8, 19). Inasmuch as oxysterols are known to increase the delivery of cholesterol from the plasma membrane to the ER (20, 21), this effect of exogenous oxysterols could be indirect and mediated via an increase in the cholesterol content of ER membranes. In a previous study using the trypsin cleavage assay, oxysterols applied to the ER membrane in vitro did not affect the conformation of SCAP (7). Because Insig expression lowers the concentration of oxysterol required to cause ER retention of SCAP/SREBP complex in intact cells (8, 9), the question arose as to whether Insig might render SCAP sensitive to oxysterols when they are applied directly to the ER membrane in vitro. We found that SCAP conformation remained insensitive to oxysterols in the presence of Insig under conditions in which Insig enhanced sensitivity to cholesterol. Whether oxysterols act in vivo by translocating cholesterol to the ER, or whether oxysterols act directly on SCAP through an intermediate protein that is missing or inactive in our in vitro assay, is currently unknown.
The third conclusion of the current studies relates to the effect of tricyclic cationic amphiphiles, which mimicked cholesterol in several ways. First, these compounds appeared to produce the same conformational change as cholesterol, i.e., the exposure of R503/R505 to trypsin. Although we did not measure the cleavage site directly, the size of the tryptic fragment was the same as achieved with cholesterol treatment as judged by SDS/PAGE. Second, these drug effects were enhanced by coexpression with Insig-1 and reduced by mutations in SCAP that reduce its interaction with Insig-1. Third, the effect of trifluoperazine was additive to that of cholesterol, suggesting that the two compounds might share a common mechanism.
Cationic amphiphiles partition into the lipid bilayer and exert both specific and nonspecific effects. The nonspecific effects relate to their disruption of membrane lipid organization and include alteration of membrane fluidity and shape, disordered trafficking of membrane lipids from one organelle to another, and the accumulation of phospholipid lamellar bodies (10–12, 22). However, some of the compounds most notable for these nonspecific membrane lipid effects, such as U18666A, diphenhydramine, and haloperidol, did not affect SCAP conformation in our assay.
On the other hand, cationic amphiphiles exert specific effects by interacting directly with proteins that contain hydrophobic domains. Examples of proteins whose activity is altered by cationic amphiphiles include G protein-coupled receptors, ion channels, membrane transporters, phospholipases, sphingomyelinase, sterol isomerases, and calmodulin (23–28). Specificity is indicated by the finding that the activity of these proteins is altered by some cationic amphiphiles, but not by others. SCAP appears to have a specific sensitivity to a subgroup of cationic amphiphiles, but this subgroup is not the same as the ones that affect other hydrophobic proteins. For example, the sterol C8-C7 isomerase is inhibited by low concentrations of trifluoperazine, but it is also inhibited by similarly low concentrations of U18666A and ifenprodil, compounds that had minimal effect on SCAP conformation when added as high as 100 μM (26, 27). The sensitivity pattern of SCAP is perhaps closest to that of the soluble protein calmodulin, whose activity is inhibited by trifluoperazine = chlorprothixene > chlorpromazine > clomipramine > haloperidol > promazine > imipramine > promethazine (24). The similarity is imperfect, however, because haloperidol has a significant effect on calmodulin (IC50 = 65 μM), but a minimal effect on SCAP when tested at 100 μM.
In our assay of SCAP conformation, the most potent nonsterol compounds possess a hydrophobic tricyclic ring structure and a positively charged amine group that is separated from the tricyclic structure by at least three atoms (Fig. 5). Hydrophobicity is likely an important determinant of potency, and the ability of a halogen group on the ring structure to increase potency may relate to the fact that halogenated tricyclic compounds are more hydrophobic than nonhalogenated ones (24). The geometry of the tricyclic ring structure is perhaps less important than its hydrophobicity, because the conformational change of SCAP could be induced by phenothiazine, dibenzazepine, or thioxanthene compounds.
The cationic amine-containing side chain was also essential for activity, raising the possibility that the effect of these compounds may be related in some way to the effect of PE in insect cells. PE is a zwitterion at physiologic pH, but it does contain a positively charged NH2 group. When present in membranes, PE and cationic amphiphiles both preferentially localize in the cytoplasmic leaflet of the plasma membrane's lipid bilayer, owing to interaction with negatively charged phosphatidylserine, which is concentrated in this leaflet (10, 29). We have been unable to obtain any alteration of SCAP conformation by addition of PE to Chinese hamster ovary cell membrane vesicles, but this is likely a technical problem, relating to the difficulty of introducing new phospholipids into existing membrane vesicles.
The notion that SCAP may monitor the physical properties of the cytoplasmic leaflet of the ER membrane is consistent with the observation that the three amino acid substitutions that reduce sterol-mediated regulation all occur in residues that would be expected to be located in the cytoplasmic leaflet. Moreover, the trypsin cleavage assay measures the exposure of an arginine residue in SCAP that lies at the interface between the cytoplasmic leaflet and the cytosol. It should be noted that the cytoplasmic leaflet of ER-derived membrane vesicles is the outer leaflet, whereas in the cell this leaflet is usually designated as the inner leaflet.
We previously proposed a model of the effect of cholesterol on SCAP conformation in which R503/R505 moves away from the cytoplasmic surface of the membrane in response to cholesterol, allowing it to be cleaved by trypsin (7). Perhaps the simplest model to explain the effect of cationic amphiphiles places them in the cytoplasmic leaflet of the vesicles, in the vicinity of SCAP. By virtue of their positive charge, the compounds repel R503/R505 away from the membrane, allowing the lower band to be generated by trypsin treatment.
Although several cationic amphiphiles mimic the effects of cholesterol on the conformation of SCAP in vitro, we have not been able to show that these compounds block the transport of the SCAP/SREBP complex from ER to Golgi in intact cells. It is possible that this failure results from an inability of the added compounds to reach the cytoplasmic leaflet of the ER membrane in intact cells. By the same token, cholesterol itself is a very poor regulator of SCAP transport when added to cells in solvents, likely because this cholesterol also fails to reach the ER membrane. Further studies of the chemical and physical properties of ER membranes will be necessary to fully explain the differences between the in vivo and in vitro assays.
Supplementary Material
Acknowledgments
We thank our colleagues Arun Radhakrishnan, Liping Sun, and Andrew Brown for helpful discussions; Arun Radhakrishnan for critical review of the manuscript; Debra Morgan for excellent technical assistance; and Lisa Beatty and Angela Carroll for invaluable help with tissue culture. This work was supported by grants from the National Institutes of Health (HL-20948), the Perot Family Foundation, and the Keck Foundation. C.M.A. is supported by the Physician Scientist Training Program at the University of Texas Southwestern Medical Center.
Abbreviations: CMV, cytomegalovirus; ER, endoplasmic reticulum; MCD, methyl-β-cyclodextrin; PE, phosphatidylethanolamine; SREBP, sterol regulatory element-binding protein; SCAP, SREBP cleavage-activating protein; TM, transmembrane segment.
References
- 1.Horton, J. D., Goldstein, J. L. & Brown, M. S. (2002) J. Clin. Invest. 109, 1125–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brown, M. S. & Goldstein, J. L. (1997) Cell 89, 331–340. [DOI] [PubMed] [Google Scholar]
- 3.Goldstein, J. L., Rawson, R. B. & Brown, M. S. (2002) Arch. Biochem. Biophys. 397, 139–148. [DOI] [PubMed] [Google Scholar]
- 4.Hua, X., Sakai, J., Brown, M. S. & Goldstein, J. L. (1996) J. Biol. Chem. 271, 10379–10384. [DOI] [PubMed] [Google Scholar]
- 5.Nohturfft, A., Brown, M. S. & Goldstein, J. L. (1998) Proc. Natl. Acad. Sci. USA 95, 12848–12853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yabe, D., Xia, Z.-P., Adams, C. M. & Rawson, R. B. (2002) Proc. Natl. Acad. Sci. USA 99, 16672–16677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brown, A. J., Sun, L., Feramisco, J. D., Brown, M. S. & Goldstein, J. L. (2002) Mol. Cell 10, 237–245. [DOI] [PubMed] [Google Scholar]
- 8.Yang, T., Espenshade, P. J., Wright, M. E., Yabe, D., Gong, Y., Aebersold, R., Goldstein, J. L. & Brown, M. S. (2002) Cell 110, 489–500. [DOI] [PubMed] [Google Scholar]
- 9.Yabe, D., Brown, M. S. & Goldstein, J. L. (2002) Proc. Natl. Acad. Sci. USA 99, 12753–12758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sheetz, M. P. & Singer, S. J. (1974) Proc. Natl. Acad. Sci. USA 71, 4457–4461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Underwood, K. W., Andemariam, B., McWilliams, G. L. & Liscum, L. (1996) J. Lipid Res. 37, 1556–1568. [PubMed] [Google Scholar]
- 12.Lange, Y. & Steck, T. L. (1994) J. Biol. Chem. 269, 29371–29374. [PubMed] [Google Scholar]
- 13.Rawson, R. B., DeBose-Boyd, R. A., Goldstein, J. L. & Brown, M. S. (1999) J. Biol. Chem. 274, 28549–28556. [DOI] [PubMed] [Google Scholar]
- 14.Seegmiller, A. C., Dobrosotskaya, I., Goldstein, J. L., Ho, Y. K., Brown, M. S. & Rawson, R. B. (2002) Dev. Cell 2, 229–238. [DOI] [PubMed] [Google Scholar]
- 15.Dobrosotskaya, I., Seegmiller, A. C., Brown, M. S., Goldstein, J. L. & Rawson, R. B. (2002) Science 296, 879–883. [DOI] [PubMed] [Google Scholar]
- 16.Schreier, S., Malheiros, S. V. P. & de Paula, E. (2000) Biochim. Biophys. Acta 1508, 210–234. [DOI] [PubMed] [Google Scholar]
- 17.Zachowski, A. & Durand, P. (1988) Biochim. Biophys. Acta 937, 411–416. [DOI] [PubMed] [Google Scholar]
- 18.Ahyayauch, H., Requero, M. A., Alonso, A., Bennouna, M. & Goni, F. M. (2002) J. Colloid Interface Sci. 256, 284–289. [DOI] [PubMed] [Google Scholar]
- 19.Wang, X., Sato, R., Brown, M. S., Hua, X. & Goldstein, J. L. (1994) Cell 77, 53–62. [DOI] [PubMed] [Google Scholar]
- 20.Brown, M. S., Dana, S. E. & Goldstein, J. L. (1975) Proc. Natl. Acad. Sci. USA 72, 2925–2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lange, Y. & Steck, T. L. (1997) J. Biol. Chem. 272, 13103–13108. [DOI] [PubMed] [Google Scholar]
- 22.Reasor, M. J. & Kacew, S. (2001) Exp. Biol. Med. 226, 825–830. [DOI] [PubMed] [Google Scholar]
- 23.Oswald, R. & Changeux, J.-P. (1981) Proc. Natl. Acad. Sci. USA 78, 3925–3929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Prozialeck, W. C. & Weiss, B. (1982) J. Pharmacol. Exp. Ther. 222, 509–516. [PubMed] [Google Scholar]
- 25.Gupta, A. K. & Rudney, H. (1991) J. Lipid Res. 32, 125–136. [PubMed] [Google Scholar]
- 26.Moebius, F. F., Striessnig, J. & Glossmann, H. (1997) Trends Pharmacol. Sci. 18, 67–70. [DOI] [PubMed] [Google Scholar]
- 27.Bae, S.-H., Seong, J. & Paik, Y.-K. (2001) Biochem. J. 353, 689–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hardman, J. G. & Limbird, L. E. (2001) Goodman and Gilman's The Pharmacological Basis of Therapeutics (McGraw–Hill, New York).
- 29.Rothman, J. E. & Lenard, J. (1977) Science 195, 743–753. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.