Abstract
Evidence is presented indicating that initiation of glycogen synthesis in Agrobacterium tumefaciens does not require the presence of α(1,4)-linked glucans. Crude cell extracts incubated with ADP-glucose (Glc) were able to form α(1,4)-linked glucans despite the fact that cells used for extract preparation displayed a genotype that prevented synthesis of Glc-containing sugar nucleotides and thus preformation of α(1,4)-linked glucans and that the defined growth medium used contained glycerol as carbon source. A. tumefaciens glycogen synthase (GS) purified to homogeneity from the above-mentioned cells was able to build its own primer by transferring Glc residues from ADP-Glc to an amino acid(s) in the same protein. Primed GS then became the substrate for further GS-catalyzed glucan elongation. It was concluded that, contrary to what happens in mammalian and yeast cells in which two different proteins are required for linear α(1,4)-linked glucan formation (glycogenin for initiation and GS for further elongation), in A. tumefaciens and probably in all other bacteria, the same protein is involved in both glycogen initiation and elongation.
Glycogen and starch synthesis involves elongation of α(1,4)-linked glucans by glycogen synthase (GS), an enzyme that uses UDP-glucose (Glc) or ADP-Glc as substrate donor in mammalian and fungal or in plant and bacterial cells, respectively (1). Polymers thus generated are substrates of an enzyme that introduces α(1,6) branches to yield the characteristic glycogen and amylopectin structures. Besides the last compound, starch also contains amylose, a linear α(1,4)-linked glucan (1).
It has been established that another enzyme, glycogenin, participates in the initiation of yeast and mammalian cell glycogen synthesis (2, 3). Glycogenin catalyzes glucosylation of a Tyr residue in the same protein by using the same sugar donors as that used by GS for further chain elongation. Glycogenin-linked maltooctasaccharides thus formed are used as primers by GS, and apparently glycogenin remains linked to the polymer through the entire elongation process (2, 3).
No glycogenin has been described in bacteria thus far, and several fully sequenced genomes of bacteria known to accumulate glycogen have failed to reveal the presence of prokaryote glycogenin homologues.
All genes involved in known Agrobacterium tumefaciens glycogen metabolism steps are clustered in a single operon formed by the sequentially transcribed glgP, glgB, glgC, glgA, pgm, and glgDB genes (4, 5). They code for glycogen phosphorylase, glycogen-branching enzyme, ADP-Glc pyrophosphorylase, GS, phosphoglucomutase, and glycogen-debranching enzyme, respectively. Although it encodes all enzymes known to be required for glycogen synthesis and utilization, the cluster does not encode a protein displaying significant similarity to glycogenin, nor does the entire A. tumefaciens genome code for such a protein (6).
In this report we present evidence indicating that A. tumefaciens GS not only elongates α(1,4)-linked glucans but also forms the primer required for the elongation process by catalyzing its own glucosylation.
Materials and Methods
Bacterial Strains and Plasmids. Escherichia coli K-12 DH5α-F′IQ and Top Ten (Invitrogen) strains were used as host for all plasmids used in this study. A. tumefaciens A348 (wild type) was provided by E. Nester (University of Washington, Seattle), A. tumefaciens mutant strains A5129 (with a Tn5 insertion in the pgm gene) and A1120 (kanamycin-resistant polar mutant in the glgB gene coding for glycogen-branching enzyme) have been described (4, 5). A. tumefaciens strain A2002 (GS null) was obtained by disrupting the glgA gene in A348 cells with a gentamicin-resistance cassette. There are two promoters in the glycogen operon: one to the 5′ end of glgP that controls transcription of the whole operon and one between the glgA and pgm genes that controls transcription of pgm and glgDB (4). Mutants A1120 and A2002 express, therefore, phosphoglucomutase and glycogen-debranching enzymes.
E. coli strains were grown at 37°C in Luria–Bertani broth (7). A. tumefaciens strains were grown at 28°C in Luria–Bertani broth or in Agrobacterium broth (AB) minimal medium with 1% glycerol as carbon source (8). If necessary, media were supplemented with the appropriate antibiotics (100 μg/ml ampicillin for E. coli or 100 μg/ml carbenicillin, 20 μg/ml gentamicin, and 50 μg/ml kanamycin for A. tumefaciens). Plasmids pBB3 and pBBR1MCS-4 have been described (4, 9).
Construction of the Double glgP-pgm Mutant. Construction of the double mutant was performed by introducing a nonpolar gentamicin-resistant cassette in the glgP gene of A5129 strain cells. Briefly, plasmid pBB3 with a 3-kb DNA fragment containing the entire glgP gene cloned in pBluescript II KS(+) was digested with PstI and ligated to a nonpolar gentamicin cassette (10). The resulting plasmid was electroporated into A5129 pgm mutant cells, and gentamicin-resistant clones were isolated. Carbenicillin-sensitive and gentamicin- and kanamycin-resistant cells represented those in which double recombination events had occurred. The genotype of selected clones was confirmed by PCR analysis by using the primers 5′-GT TGCGGT TATGGTCTGC-3′ and 5′-CGGGCAGAAGCGTGTGGT-3′, which amplify 0.4- and 1.24-kb DNA fragments in the parental and double-mutant cells, respectively. A double-mutant strain clone (A5130) was selected for further studies.
Cloning and His Tagging of the glgA Gene. To express the GS-encoding gene (glgA) with a C-terminal poly-His tag, the gene was amplified by PCR by using the primers 5′-AAGCTGGACCTGTAGAGCCC-3′ and 5′-TCAATGATGATGATGATGATGGCCTTTCGAAATAA-3′ and Pwo polymerase. The PCR product was cloned in pGemt-T Easy vector (Promega), the resulting plasmid was digested with ApaI and SpeI, and the DNA fragment was cloned in plasmid pBBR1MCS-4 previously digested with the same enzymes. The resulting plasmid, named pBG19, codes for A. tumefaciens GS displaying six His residues at its C terminus.
Determination of GS Initiation and Elongation Activities in Crude Extracts. Cells in the late exponential growth phase were harvested from 100 ml of medium and resuspended in 5 ml of 50 mM Tris·HCl buffer, pH 7.6/5 mM EDTA/20% sucrose/1 mMPMSF/200 μg/ml lysozyme. Cells were recovered by centrifugation after incubation for 2 h at 4°C, resuspended in 1 ml of 50 mM Tris·HCl buffer, pH 7.6/5% sucrose/3 mM 2-mercaptoethanol and disrupted by sonication. Supernatants obtained on 15,000 × g for 30-min centrifugations were used as crude extracts.
GS-unprimed initiation activity was estimated in incubation mixtures containing, in a total volume of 50 μl, 100 mM glycine-NaOH buffer, pH 8.7, 5 mM DTT, 100 mM EDTA, 3 μM ADP-[14C]Glc [300 μCi/μmol (1 Ci = 37 GBq)], and 25–30 μg of crude extract protein or 2.5 μg of purified GS. The labeled sugar nucleotide was synthesized as described before for UDP-[14C]Glc by using all enzymes from Saccharomyces cerevisiae except that recombinant E. coli ADP-Glc pyrophosphorylase instead of yeast UDP-Glc pyrophosphorylase was used (11). Reactions were stopped after 30 min at 30°C by the addition of 10 volumes of 10% trichloroacetic acid (TCA). Tubes were centrifuged after1hat4°C, precipitates were washed three times with 10% TCA, and label incorporated was quantified.
GS-unprimed elongation activity was estimated as described above, but the incubation mixture contained 2.5 mM ADP-[14C]Glc (0.36 μCi/μmol) (unlabeled ADP-Glc was from Sigma).
GS-primed elongation activity was estimated by label incorporated into 75% methanol/1% KCl-insoluble fraction. Reaction mixtures contained, in a total volume of 100 μl, 100 mM Mops buffer, pH 7.0, 25 mM KCl, 10 mg of rabbit muscle glycogen (Sigma), and 1.25 mM ADP-[14C]Glc (0.72 μCi/μmol). After 30 min at 30°C, reactions were stopped by the addition of 3 volumes of 75% methanol/1% KCl. Pellets were washed three times with the same solution, and the radioactivity incorporated into precipitates was quantified.
Purification of Recombinant GS. Soluble extract preparation. Recombinant GS was purified from 20 g (wt/wt) of strain A5130 cells harboring plasmid pBG19 grown in AB medium with 1% glycerol as carbon source. Cells were harvested, washed once with 35 ml of 50 mM Tris·HCl buffer, pH 8.0, and resuspended in 200 ml of 50 mM Tris·HCl buffer, pH 7.6/20% sucrose/5 mM EDTA/1 mM PMSF/200 μg/ml lysozyme and incubated for 2 h at 4°C. Cells were recovered by centrifugation, resuspended in 30 ml of 50 mM Tris·HCl buffer, pH 7.6/5% sucrose/3 mM 2-mercaptoethanol (solution A), containing protease inhibitors (1 mM tosylphenylalanyl chloromethyl ketone/1 mM Nα-p-tosyl-l-lysine chloromethyl ketone/1 mM PMSF/1 μM E-64/1 mM pepstatin/10 μM leupeptin), and 20 μg/ml DNase and disrupted by three compression–decompression cycles in a French press. The suspension was centrifuged for 30 min at 15,000 × g, and the supernatant was centrifuged for 120 min at 100,000 × g.
Ammonium sulfate precipitation. Supernatants obtained after ultra-centrifugation were salted-out with 65% saturation (NH4)2SO4, and pellets were dissolved in 50 ml of solution A and dialyzed overnight against the same buffer.
DEAE-Sepharose chromatography. The dialyzed solution was applied to a DEAE-Sepharose column (1.5 × 15 cm) equilibrated with solution A. Elution was performed with a linear 0–0.6 M NaCl gradient (≈5 column volumes). Fractions showing enzymatic activity (GS-primed elongation assay; active fractions contained ≈0.25 M NaCl) were pooled and dialyzed overnight against solution A without 2-mercaptoethanol at 4°C (solution B).
Chelating-agarose chromatography. The solution then was applied to a Ni2+-iminodiacetic-agarose column (0.5 × 4.0 cm; Sigma) equilibrated with solution B. The column was washed with solution B containing 0.5 M NaCl until no absorbance at 280 nm was detected. Elution was performed in one step with 15 ml of solution B containing 50 mM EDTA at a flow rate of 1 ml/min. The eluted solution was salted-out with 65% saturated (NH4)2SO4, and the pellet was dissolved in 20 ml of solution A and dialyzed overnight against the same buffer.
Mono Q chromatography. The dialyzed solution was filtered through a Millipore 0.22-μm filter and applied to a Mono Q HR 5/5 column equilibrated with solution A. Two 25-ml linear NaCl gradients (0–0.05 and 0.05–0.25 M) in solution A were sequentially applied to the column. The flux was 1 ml/min in both gradients.
Phenyl-Superose chromatography. Active fractions recovered from the Mono Q column chromatography were pooled, diluted 10-fold with solution A containing 1.5 M (NH4)2SO4 (solution C), and applied to a phenyl-Superose HR 5/5 column equilibrated with the same solution. The enzyme was eluted with a 12.5-ml linear solution C–solution A gradient (flux 0.5 ml/min). Active fractions were pooled. The final preparation displayed a single protein band when run in 10% SDS/PAGE and further stained with Coomassie brilliant blue. Hydrophobic chromatography or both Mono Q and hydrophobic chromatographies were repeated once if more than one band appeared.
Construction of Plasmid Encoding S. cerevisiae GSII. The GlgCII gene from S. cerevisiae was amplified by using Pwo polymerase, genomic DNA as template, and the primers 5′-CCAAGCTTAGAGCCCATGTCCCGTGACCTACA-3′, which has a HindIII restriction site and a Shine–Dalgarno sequence upstream to the start codon, and 5′-CGGGATCCATTTAACTGTCATCAGCATA-3′, which has a BamHI restriction site and includes the stop codon. The PCR product was digested with BamHI/HindIII and ligated to plasmid pBBR1MCS-4 previously digested with the same enzymes. The resulting plasmid was designated pGSCII and introduced into A1120 cells by triparental mating.
Proteinase K and α-Amylase Treatments. Unprimed initiation reactions were heat-inactivated at 65°C for 15 min, after which 5 μg of α-amylase (Sigma) or 3 μg of proteinase K (BRL and Life Technologies, Rockville, MD) were added. Incubations with α-amylase lasted for 2 h at 20°C, and those with proteinase K lasted for 1 h at 65°C. Reactions were stopped by the addition of 10 volumes of 10% TCA, and pellets were submitted to 10% SDS/PAGE followed by autoradiography.
Additional Methods. Glycogen was quantified as described by Krisman (12) with oyster glycogen (Sigma) as standard, but absorbance at 510 nm and not at 450 nm was determined to circumvent unspecific absorption. Glycogen in bacterial colonies was detected by exposing them to iodine vapors.
Results
De Novo Bacterial Glycogen Synthesis Does Not Require the Presence of an α(1,4)-Linked Glycan Acceptor Primer. To determine whether an α(1,4)-linked glucan was absolutely required as primer for glycogen synthesis, we constructed A. tumefaciens double mutant glgP-pgm (A5130). These strain cells lack α(1,4)-linked glucan phosphorylase and thus are unable to form Glc 1-phosphate by phosphorolysis of α(1,4)-linked glucans; because they are also devoid of phosphoglucomutase, they cannot synthesize either ADP-Glc or UDP-Glc from Glc 6-phosphate because they cannot transform this last compound in Glc 1-phosphate. Absolutely no glycogen was detected in mutant cells grown to late stationary phase in AB medium containing 1 mM ammonium chloride and 1% sucrose as carbon source, that is, under optimal conditions for glycogen accumulation. Wild-type cells had 3 mg of glycogen per g of wet weight under similar conditions.
Crude extracts prepared from A5130 cells grown in a defined, minimal medium with glycerol as carbon source incubated with a low ADP-[14C]Glc concentration (unprimed initiation conditions) synthesized ≈14-fold more of a 10% TCA-insoluble product than a similar extract prepared from wild-type cells (Table 1). In contrast, the wild-type cell extract synthesized >100-fold more of the insoluble product when ADP-Glc concentration was raised from 3 μM to 2.5 mM (unprimed elongation conditions) (Table 1). This result was not due to significantly different GS levels in both extracts because that from wild-type cells only showed a 2.7-fold higher label incorporation into glycogen when this polysaccharide was added to incubation mixtures (primed elongation conditions) (Table 1). On the other hand, as observed in the Table 1, no incorporation was observed under any of the experimental conditions used when the crude extract was prepared from A1120 (branching enzyme, ADP-Glc pyrophosphorylase, and GS null) mutant cells. As will be described below, the 10% TCA-insoluble labeled product obtained when A5130 cell extracts were used was identified as an α(1,4)-linked glucan.
Table 1. Label incorporation by crude extracts.
| Strains
|
|||
|---|---|---|---|
| Incubation conditions | Wild type (A348) | glgP/pgm (A5130) | glgB polar (A1120) |
| Unprimed initiation | 0.0038 | 0.053 | ND |
| Unprimed elongation | 85 | 0.77 | ND |
| Primed elongation | 717 | 262 | ND |
Reactions were carried out as described in Materials and Methods by using crude extracts as enzyme source. Activities are expressed as nmol of glucose·h-1 per mg of protein. ND, not detected.
Because results obtained under both unprimed incubation conditions were reminiscent of what happens in eukaryotic cells in which glycogenin (glycogen initiation) shows a much lower Km for the sugar nucleotide than GS (glycogen elongation) (2), it was tentatively concluded that two different reactions (glucan initiation and elongation) occurred in our experimental system. Higher formation of the insoluble product under unprimed initiation conditions in A5130 cell extracts may be ascribed to higher amounts of unoccupied sites in a protein primer, whereas preformed protein-linked α(1,4)-glucans in wild-type cell extracts may account for the higher incorporation observed in them under unprimed elongation conditions. Results obtained demonstrate that, the same as in eukaryotic cells, bacterial glycogen de novo synthesis does not require the presence of α(1,4)-linked glucans.
S. cerevisiae GS Expressed in A. tumefaciens Participates in Elongation but Not in Initiation Reactions. It has been reported that disruption of both S. cerevisiae glycogenin-encoding genes completely abolished glycogen synthesis in the live yeast, thus indicating that S. cerevisiae GS is unable to initiate glycogen synthesis (13). An expression vector coding for S. cerevisiae GSII was introduced into A. tumefaciens A1120 strain, which lacks glycogen-branching enzyme, ADP-Glc pyrophosphorylase, and GS activities. As a control, an expression vector coding for A. tumefaciens GS was also introduced into A1120 strain cells. Results obtained in unprimed initiation and primed elongation assays performed by using crude extracts prepared from the parental (A1120) as well as the S. cerevisiae GSII-expressing (A1120, pGSCII) and the A. tumefaciens GS-expressing (A1120, pBG19) cells are depicted in Table 2. It may be observed that in the absence of glycogen the bacterial but not the yeast GS was capable of initiating α(1,4)-linked glucan synthesis. In contrast, both extracts yielded almost similar label incorporations in the presence of the exogenous polysaccharide. It is worth mentioning that both GSs were in a similar plasmid under the control of the same promoter. Results shown in Table 2 confirm, therefore, the absence of an α(1,4)-linked glucan primer in the bacterial extracts used.
Table 2. Glucan synthesis by S. cerevisiae and A. tumefaciens GSs.
| Incubation conditions
|
||
|---|---|---|
| Strain | Unprimed initiation | Primed elongation |
| A1120 | ND | ND |
| A1120 (pGSCII) | ND | 3,284 |
| A1120 (pBG19) | 0.26 | 5,829 |
Reactions were carried out as described in Materials and Methods by using crude extracts as enzyme source. Activities are expressed as nmol of glucose·h-1 per mg of protein. ND, not detected.
An in vivo complementation assay was performed by introducing plasmids pBG19 (A. tumefaciens GS) or pGSCII (S. cerevisiae GS) into GS null mutant cells (A2002). Resulting strains were grown on plates in AB medium with 1% sucrose as carbon source and exposed to iodine vapors for glycogen detection. Only transformation with pBG19 resulted in glycogen accumulation, thus confirming that bacterial but not yeast GS was able to synthesize glycogen de novo.
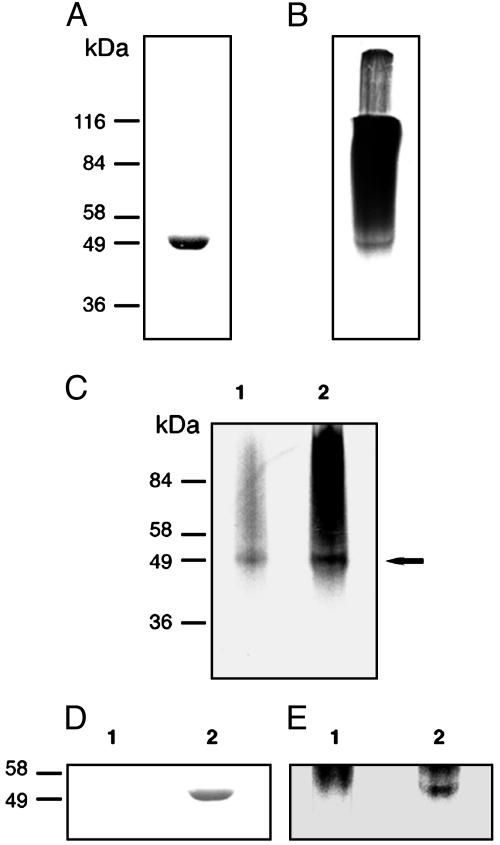
Purified A. tumefaciens GS Synthesizes Glycogen de Novo. To study the possibility that GS could be acting both as primer and elongating enzyme in bacteria, a recombinant A. tumefaciens GS containing a poly-His tag at its C terminus was expressed in A. tumefaciens A5130 mutant cells grown in minimal medium with glycerol as carbon source. The recombinant protein was purified to homogeneity and submitted to SDS/PAGE. As depicted in Fig. 1A, a single band displaying the molecular mass expected for GS (50 kDa) appeared on staining gels with Coomassie brilliant blue even after overloading gels.
Fig. 1.
SDS/PAGE of A. tumefaciens GS-reaction products. Purified GS was incubated with ADP-[14C]Glc under unprimed initiation reaction conditions, and 10% TCA-insoluble material was submitted to 10% SDS/PAGE. (A) Coomassie brilliant blue staining. (B) Autoradiography of the gel shown in A.(C) Reactions were carried out as described for A and stopped by heating tubes at 65°C for 15 min, the contents were treated with α-amylase, and precipitates insoluble in 10% TCA were submitted to 10% SDS/PAGE. Lane 1, enzymatically treated; lane 2, untreated control. The arrow indicates GS migrating position. (D and E) Reactions were carried out and stopped as described for C and submitted to proteinase K treatment, and precipitates insoluble in 10% TCA were run on 10% SDS/PAGE. (D) Coomassie brilliant blue staining. (E) Autoradiography of the gel shown in D. Lanes 1, enzymatically treated samples; lanes 2, untreated controls. For further details see Materials and Methods.
Unprimed initiation and elongation reactions performed with the purified enzyme yielded label incorporations of 8 and 11,520 nmol of Glc·h–1 per mg of protein, respectively. These results indicate that bacterial GS is capable by itself of carrying out de novo α(1,4)-linked glucan synthesis in the absence of a glucan primer.
Identification of a GS-Glucosylated Form. Material insoluble in 10% TCA yielded by purified GS incubated under unprimed initiation conditions was run in SDS/PAGE, and gels were submitted to autoradiography. As depicted in Fig. 1B, three differently labeled fractions could be detected in the gel: the first one entered into the stacking but not into the separating gel; the second one entered into the latter; and finally the third fraction migrated exactly as GS (Fig. 1 A and B). Label in gels drastically diminished when reactions were stopped by heating tubes at 65°C for 15 min and the resulting solutions were treated with α-amylase before being submitted to SDS/PAGE (Fig. 1C, lanes 1 and 2) (although not shown, label in the stacking gel also diminished). This result indicates that label in the three above-mentioned fractions was in α(1,4)-linked glucans. To confirm that the faster-migrating fraction indeed was a protein-linked glucan, the heated reaction mixture was treated with proteinase K before SDS/PAGE. As depicted in Fig. 1 D and E, both the Coomassie brilliant blue-stained and the labeled 50-kDa bands disappeared after the proteolytic treatment.
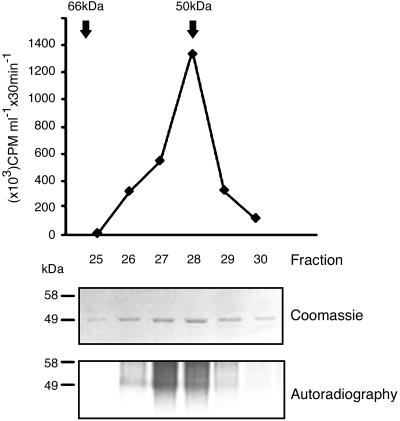
To establish whether the faster-migrating fraction in Fig. 1B represented label incorporation into GS and not into a contaminating protein having the same molecular mass as GS in SDS/PAGE and being intimately associated with it under non-denaturing conditions, purified GS was submitted to gel filtration through a BioSil SEC 250 (Bio-Rad) column, and incorporation under unprimed initiation conditions was assayed in resulting fractions. As depicted in Fig. 2, the active protein displayed the same molecular mass as GS (50 kDa), thus confirming that GS itself had been labeled in the assays. The active fractions would have been expected to display a molecular mass of at least 100 kDa in the BioSil column if label had been incorporated into a 50-kDa protein that had remained GS-associated during all purification procedures of the latter.
Fig. 2.
GS gel filtration. Purified GS was submitted to BioSil SEC 250 gel chromatography. The arrows indicate the elution volumes of 66- and 50-kDa protein standards. Fractions were submitted to 10% SDS/PAGE and stained with Coomassie brilliant blue. The same fractions were used as enzyme source in unprimed initiation reactions. TCA precipitates were submitted to 10% SDS/PAGE followed by autoradiography. For further details see Materials and Methods.
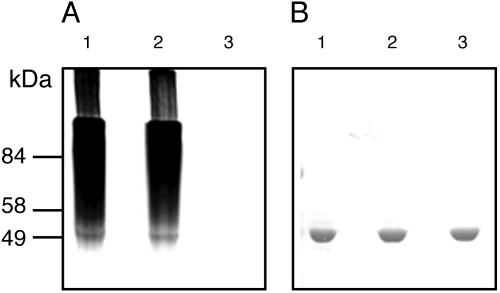
GS-Linked Glucans Are Intermediaries in the Synthesis of Large Polymers. Purified GS labeling after incubation with ADP-[14C]Glc revealed that initiation proceeded without the participation of any other protein. To confirm that indeed labeled GS was an intermediate in the synthesis of large polymers, purified GS was incubated under unprimed initiation conditions (3 μM ADP-[14C]Glc) for 15 or 30 min or alternatively for only 15 min, after which unlabeled ADP-Glc to a final 5 mM concentration was added and the sample was incubated further for 15 min. Material insoluble in 10% TCA was submitted to 10% SDS/PAGE followed by autoradiography or Coomassie brilliant blue staining. As depicted in Fig. 3A, label disappeared from the pulse–chased sample, thus indicating that a large polymer, unable to enter even into the stacking gel, had been formed during the chase. Unexpectedly, staining did not reveal a discernable decrease in the amount of free GS (Fig. 3B). Further, we failed to observe a decrease in the amount of GS when only minute amounts of it and high unlabeled ADP-Glc concentrations were used. This indicates that either only minimal amounts of GS had been used as primer or, alternatively, that GS had been released from the polymer at some point during the elongation process. We favor the first interpretation, because, as Figs. 1 B and C and 3A show, under unprimed initiation conditions large polymers were formed that nevertheless entered into the stacking and separating gels, thus indicating that the α(1,4)-linked glucans formed were still linked to a protein moiety. Protein-linked but not protein-free glycans enter into gels (14).
Fig. 3.
GS-linked glucans are intermediates in the synthesis of large polymers. Purified GS was incubated under unprimed initiation conditions for 15 or 30 min with 3 μM ADP-[14C]Glc (lanes 1 and 2, respectively) or for only 15 min, after which unlabeled ADP-Glc toa5mM final concentration was added, and the sample was incubated further for 15 min (lanes 3). Material insoluble in 10% TCA was submitted to 10% SDS/PAGE followed by autoradiography (A) or staining with Coomassie brilliant blue (B). For further details see Materials and Methods.
Discussion
Based on the formation of labeled 10% TCA-insoluble products yielded by incubation mixtures containing as enzyme source a pellet of 105,000 × g for 150 min centrifugations of whole E. coli cell extracts and either UDP-[14C]Glc or ADP-[14C]Glc, it was suggested that both sugar nucleotides could serve as sugar donors in the glucosylation of a protein primer that further ADP-Glc-dependent elongation and branching converted into bacterial high molecular weight glycogen (15, 16). This suggestion was later questioned, because it was demonstrated that not only the cell pellets used in the assays but also homogenous E. coli GS preparations that were also able to synthesize α(1,4)-linked glucans on incubations with labeled sugar nucleotides contained noncovalently bound glucans that could serve as primers for polymer elongation (14, 17). It is a well known fact that GSs tightly associate with α(1,4)-linked glucans, and this property has been widely used to purify GSs from several sources. Further, it was demonstrated that insolubility in 10% TCA did not necessarily imply that a glucan was covalently linked to a protein as protein-free linear α(1,4)-linked glucans (that is, those synthesized in vitro in the presence of maltooligosaccharides as primers and in the absence of branching enzyme) were shown to be intrinsically insoluble in acidic media (14).
To overcome possible contamination with α(1,4)-linked glucans we used A. tumefaciens cells unable to synthesize Glc 1-phosphate and thence also any Glc-containing sugar nucleotide because they lacked both α(1,4)-glucan phosphorylase and phosphoglucomutase activities. Moreover, we used a minimal growth medium with glycerol as carbon source. A somewhat similar approach has been used to synthesize Glc-free recombinant muscle glycogenin in E. coli. In this case, because glycogenin was already known to specifically use UDP-Glc as sugar donor, the mutation introduced in the bacterial strain abolished UDP-Glc pyrophosphorylase activity (18). The absence of α(1,4)-linked glucans in A. tumefaciens cells used here was confirmed not only by the absence of glycogen under growth conditions optimal for the accumulation of the polymer but also by the inability of S. cerevisiae GS (an enzyme absolutely dependent on α(1,4)-glucan primers for Glc transfer reactions) to synthesize those same glucans both in in vitro and in vivo assays when expressed in A. tumefaciens mutant cells. We recently crystallized recombinant A. tumefaciens GS purified as described herein and solved its structure to a 2.3-Å resolution (unpublished data; see a preliminary report in ref. 19). The structural study confirmed that the enzyme was devoid of covalently linked or intimately associated glycans.
Incubation of homogeneous GS preparations derived from A. tumefaciens cells unable to form Glc-containing sugar nucleotides with ADP-[14C]Glc led to formation of both labeled molecules that migrated the same as the pure enzyme and of larger polymers. Although the former signal certainly corresponded to [14C]Glc covalently linked to a protein as it disappeared after a proteolytic treatment, the larger compounds were also covalently linked to protein as they entered into the stacking and separating gels in SDS/PAGE. Probably large glucans prevented proteinase K from fully degrading GS, because the larger compounds entered into the gels even after the proteolytic treatment. Glucans migrating either as GS or displaying larger sizes were degraded by α-amylase, thus showing that they were α(1,4)-linked polymers. Primed GS then became the substrate for further GS-catalyzed glucan elongation. The preferential utilization of GS not as primer but as elongator observed might be explained by the above-mentioned tight association between GS molecules and large α(1,4)-linked glucans. That is, once a relatively large polymer is formed, GS would be committed mainly to elongation of branches with which it strongly interacts.
Whereas in yeast and mammalian cells two different proteins are required for linear α(1,4)-linked glucan formation (glycogenin for initiation and GS for further elongation) (2, 13), A. tumefaciens GS seems to play both roles. This conclusion is probably valid for all eubacteria because of the high similarity found among different species of GS primary sequences. It has not been firmly established yet whether starch synthesis in plants (a process that uses ADP-Glc as sugar donor, the same as glycogen synthesis in bacteria) follows the mammalian/yeast or bacterial models. It has been reported that homogeneous recombinant maize starch synthase I was able to synthesize an α(1,4)-linked glucan when incubated with ADP-Glc (20). Although the enzyme had been synthesized in an E. coli strain fully capable of glycogen synthesis, it contained <1 Glc residue per 26 GS molecules. On the other hand, eight genes coding for proteins showing significant similarity to mammalian cell glycogenins (in all cases <40%) have been detected in the Arabidopsis thaliana genome. Whether any of the encoded proteins display the known autoglucosylating capacity of glycogenins or whether, alternatively, all of them represent other members of the glycosyltransferase family 8 is presently unknown.
Genes coding for a number of anabolic and catabolic pathways are clustered in operons in prokaryotes to guarantee their coordinately regulated expression. As mentioned above, in the case of A. tumefaciens not only genes involved in glycogen synthesis but also in its degradation seem to be part of the same operon. Our finding reported herein that GS behaves not only as an elongator but also as an initiator is consistent with the clustered organization of glycogen genes.
Acknowledgments
The technical skills of Susana Raffo in the synthesis of ADP-[14C]Glc and of Andrea Merás in protein purification are gratefully acknowledged. This research was supported by grants from the National Agency for the Promotion of Science and Technology (Argentina) and the Howard Hughes Medical Institute. J.E.U. is a doctoral fellow of the National Research Council (Argentina), and R.A.U. and A.J.P. are Career Investigators of the same institution.
Abbreviations: GS, glycogen synthase; Glc, glucose; TCA, trichloroacetic acid.
References
- 1.Preiss, J. & Romeo, T. (1989) Adv. Microb. Physiol. 30, 183–238. [DOI] [PubMed] [Google Scholar]
- 2.Alonso, M. D., Lomako, J., Lomako, W. M. & Whelan, W. J. (1995) FASEB J. 9, 1126–1137. [DOI] [PubMed] [Google Scholar]
- 3.Smythe, C. & Cohen, P. (1991) Eur. J. Biochem. 200, 625–631. [DOI] [PubMed] [Google Scholar]
- 4.Ugalde, J. E., Lepek, V., Uttaro, A., Estrella, J., Iglesias, A. & Ugalde, R. A. (1998) J. Bacteriol. 180, 6557–6564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Uttaro, A. D. & Ugalde, R. A. (1994) Gene 150, 117–122. [DOI] [PubMed] [Google Scholar]
- 6.Wood, D. W., Setubal, J. C., Kaul, R., Monds, D. E., Kitajima, J. P., Okura, V. K., Zhou, Y., Chen, L., Wood, G. E., Almeida, N. F., Jr., et al. (2001) Science 294, 2317–2323. [DOI] [PubMed] [Google Scholar]
- 7.Sambrook, J., Fritsch, E. F. & Maniatis, T. (1989) Molecular Cloning: A Laboratory Manual (Cold Spring Harbor Lab. Press, Plainview, NY).
- 8.Douglas, C. J., Staneloni, R. J., Rubin, R. A. & Nester, E. W. (1985) J. Bacteriol. 161, 850–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kovach, M. E., Elzer, P. H., Hill, D. S., Robertson, G. T., Farris, M. A., Roop, R. M., 2nd, & Peterson, K. M. (1995) Gene 166, 175–176. [DOI] [PubMed] [Google Scholar]
- 10.Ugalde, J. E., Czibener, C., Feldman, M. F. & Ugalde, R. A. (2000) Infect. Immun. 68, 5716–5723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wright, A. & Robbins, P. W. (1965) Biochim. Biophys. Acta 104, 594–595. [Google Scholar]
- 12.Krisman, C. R. (1962) Anal. Biochem. 4, 17–23. [DOI] [PubMed] [Google Scholar]
- 13.Cheng, C., Mu, J., Farkas, I., Huang, D., Goebl, M. G. & Roach, P. J. (1995) Mol. Cell. Biol. 15, 6632–6640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Holmes, E. & Preiss, J. (1979) Arch. Biochem. Biophys. 196, 436–448. [DOI] [PubMed] [Google Scholar]
- 15.Barengo, R., Flavia, M. & Krisman, C. R. (1975) FEBS Lett. 53, 274–278. [DOI] [PubMed] [Google Scholar]
- 16.Barengo, R. & Krisman, C. R. (1978) Biochim. Biophys. Acta 540, 190–196. [DOI] [PubMed] [Google Scholar]
- 17.Kawaguchi, K., Fox, J., Holmes, E., Boyer, C. & Preiss, J. (1978) Arch. Biochem. Biophys. 190, 385–397. [DOI] [PubMed] [Google Scholar]
- 18.Alonso, M. D., Lomako, J., Lomako, W. M., Whelan, W. J. & Preiss, J. (1994) FEBS Lett. 352, 222–226. [DOI] [PubMed] [Google Scholar]
- 19.Guerin, M. E., Buschiazzo, A., Ugalde, J. E., Ugalde, R. A. & Alzari, P. M. (2003) Acta Crystallogr. D 59, 526–528. [DOI] [PubMed] [Google Scholar]
- 20.Imparl-Radosevich, J. M., Li, P., Zhang, L., McKean, A. L., Keeling, P. L. & Guan, H. (1998) Arch. Biochem. Biophys. 353, 64–72. [DOI] [PubMed] [Google Scholar]