Abstract
The BtuB transporter mediates high-affinity binding and TonB-dependent active transport of vitamin B12 [cyanocobalamin (CNCbl)] across the outer membrane of Escherichia coli. A characteristic feature of TonB-dependent transporters is the Ton box, a conserved sequence near the N terminus and exposed to the periplasm. Crosslinking to TonB and site-directed spin labeling indicated that the Ton box of BtuB undergoes a substantial conformational transition in response to CNCbl binding, but only slight movement was seen in crystal structures. An in vivo method of detecting substrate-induced changes in the Ton box environment measured reaction of a biotin maleimide derivative with cysteine substitutions through the N-terminal region of BtuB between positions 1 and 31. The degree of maleimide labeling of different residues correlated with their accessibility in the crystal structure. Labeling of many positions was increased strongly when CNCbl was present, consistent with the undocking of this region proposed from spin-labeling analyses. The receptor-binding domain of colicin E3, which binds to BtuB competitively with CNCbl, resulted in decreased labeling. Both substrate-induced transitions occur in and beyond the Ton box and were affected by transport-uncoupling substitutions. Thus, two transport substrates that bind competitively to the extracellular face of BtuB stabilize opposite transitions of the Ton box.
Keywords: vitamin B12, colicins, outer membrane, transmembrane signaling, transporter structure
The TonB-dependent transporters in the outer membrane (OM) of Escherichia coli and other Gram-negative bacteria mediate the active transport into the periplasm of cobalamins (Cbls) such as vitamin B12 [cyano-Cbl (CNCbl)] and of iron complexed to siderophores or heme or released from iron-binding proteins. Transport activity is coupled to the proton-motive force by the transperiplasmic protein TonB and the cytoplasmic membrane proteins ExbB and ExbD (see recent reviews in refs. 1–3). The structures of the E. coli TonB-dependent transporter for iron-siderophores FhuA (4, 5), FepA (6), and FecA (7) as well as the CNCbl transporter BtuB (8) show similar 22-stranded β-barrels enclosing a 135- to 150-residue N-terminal domain called the cork, plug, or hatch. The substrate-binding surfaces on the extracellular side of these proteins are formed from apical loops of the hatch domain and some external loops of the barrel.
TonB-dependent transporters are also receptors for lethal agents including phages, microcins, and colicins. FepA and FhuA are receptors for some group B colicins, the entry of which requires the TonB system. BtuB is receptor for the A and E colicins, the entry of which requires instead the Tol system (reviewed in refs. 9 and 10). Colicins are elongated proteins, the T, R, and C domains of which mediate OM translocation, receptor binding, and lethal activity, respectively. Binding of E colicins to BtuB is competitively inhibited by CNCbl (11). The 76-aa receptor-binding R domain of colicin E9 binds to BtuB with similar high affinity as the intact colicin (12). Competition between CNCbl and colicin could reflect their binding to the same transporter surfaces even though their uptake follows different routes.
The mechanism by which TonB promotes transmembrane movement and release of substrates from the transporters is unknown. TonB action is required for detectable transport activity (13) and likely promotes the structural rearrangement that unblocks the hatch region to allow substrate passage. An initial step in TonB-dependent transport is the signal transduction event in which substrate binding triggers a conformational transition on the periplasmic face, which could direct TonB to substrate-loaded transporters. A key factor in signaling and transport is the conserved sequence near the N terminus of TonB-dependent transporters called the Ton box (the sequence in BtuB is DTLVVTA, residues 6–12). This region is the site of mutations, such as the substitution of Pro for Leu-8 or Val-10 in BtuB, which result in loss of Cbl transport but do not affect Cbl binding or entry of the TonB-independent phage BF23 and E colicins (13, 14). The transport defect in Ton box mutants is partially reversed by suppressor mutations affecting nearby residues in the Ton box or residue Gln-160 of TonB (15, 16).
The nature of the change in Ton box conformation after substrate binding is not well defined. The Ton box is disordered in the structures of FhuA and FecA, but a short helix to which the Ton box is attached is unfolded in the substrate-bound structures and moves across the periplasmic opening (5, 7). Unfolding of a switch helix is not a general feature because it is absent in FepA and BtuB. Cys substitutions in the Ton box of BtuB or FecA form disulfide bonds in vivo with Cys residues in TonB around position 160, the site of suppressor mutations (17, 18). Only certain pairs of Cys residues efficiently formed disulfide bonds, suggesting a specific orientation of their approach. Most crosslinks were stimulated by CNCbl. Other TonB-dependent transporters also show increased interaction with TonB when their substrates were added, as detected by formaldehyde-mediated crosslinking or adsorption to matrix-immobilized TonB (19–21). All interactions were altered by uncoupling substitutions in the Ton box.
The EPR spectra of site-directed nitroxide spin labels provide information on local secondary structure, side-chain tertiary contacts, and backbone dynamics (22). Spin probe on some positions in the BtuB Ton box experience restricted motion, whereas adjacent positions were highly mobile (23). The addition of CNCbl resulted in high mobility at all Ton box positions, indicating that CNCbl binding releases the Ton box from contacts in BtuB such that it can extend into the periplasm to contact TonB. Surprisingly, the crystal structures of BtuB revealed only small changes in the Ton box after CNCbl binding (8).
We describe here a complementary approach to investigate substrate-induced changes in the N-terminal segment of BtuB including the Ton box. The extent of covalent coupling in vivo of the sulfhydryl reagent 1-biotinamido-4-[4′-(maleimidomethyl)-cyclohexane-carboxamido]butane (BMCC) to Cys substitutions in the N-terminal region of BtuB can reflect different structural features of side-chain accessibility than are revealed by spin labeling or crystallography. We found that BMCC labeling behavior can reconcile the apparent differences raised by the other techniques. BMCC labeling reveals decreased access of Ton box residues, consistent with their accessibility in the crystal structure (8). However, labeling showed a dramatic increase in access after CNCbl binding, matching the EPR results. Two TonB-uncoupled substitutions in BtuB had different effects on the labeling patterns of the Ton box. Strikingly, another BtuB substrate, the colicin E3 receptor-binding domain (E3R), had the opposite effect on accessibility of the Ton box from CNCbl. The different behavior of the Ton box shown in the various assays can be largely reconciled.
Experimental Procedures
Strains and Plasmids. Site-directed mutagenesis of the btuB gene by using a two-step PCR technique with single mutagenic oligonucleotides was as described (17). Sequences of mutagenic oligonucleotide primers are available on request. Mutant alleles were transferred by exchange of HindIII–BsiWI fragments into the corresponding sites of plasmid pAG1, a pUC8 derivative carrying the btuB ORF and regulatory region (14). The presence of expected sequence changes was verified by automated DNA sequencing. Plasmids were introduced by transformation into strain RK5016, a derivative of MC4100, with genotype Δ(argF-lac)U169 rpsL150 araD139 relA1 flbB5301 deoC1 ptsF25 rbsR22 non-9 gyrA219 metE70 argH btuB recA. Strain BL21λDE3 (Novagen) was used for expression of the E3R.
Expression of E3R Domain. DNA encoding the 76-aa E3R, having the same sequence as the colicin E9 domain (12), was amplified by PCR with primers that introduced an NdeI site at the 5′ end and the coding region for a His6 tag and an EcoRI site at the 3′ end. Template DNA was plasmid pColE3-CA38. This DNA fragment was ligated into expression vector pET17b (Novagen) digested with NdeI and EcoRI. The resulting plasmid pNC7 was introduced into E. coli BL21λDE3 by transformation. For protein production, cells grown to OD595 of 0.6–0.7 were induced with 0.25 mM isopropyl β-d-thiogalactoside for 3–4 h. Cells were collected by centrifugation, washed, and disrupted in a French pressure cell at 20,000 psi (1 psi = 6.89 kPa). The E3R fragment was purified from the cell lysate by adsorption to nickel-agarose matrix and elution with imidazole according to manufacturer recommendations (Qiagen, Valencia, CA). The protein fraction was dialyzed against 20 mM Tris·HCl (pH 8.0)/100 mM NaCl.
Maleimide Labeling of BtuB in Intact Cells. Plasmid pAG1, encoding Cys-less wild-type BtuB, and derivatives encoding BtuB variants with single Cys or double Pro and Cys substitutions were transferred into strain RK5016 by transformation. Overnight cultures were diluted 1:10 in salts medium A with 0.2% glucose, 0.01% methionine and arginine, and ampicillin at 100 μg/ml as described (17, 24). After growth at 37°C for 1.5 h, each culture was split into three portions. One portion received E3R at 5 μg/ml, which is sufficient to protect cells against killing by colicin E3. After 15 min, 5 μM CNCbl was added to a second portion, and biotin maleimide (EZ-Link Biotin-BMCC, Pierce Endogen) at a final concentration of 15 μg/ml was added to all three portions. Labeling proceeded for 15 min at 37°C and was stopped with 10 mM DTT. Cells were washed by centrifugation and suspended in PBS to OD595 of 0.75; 10 μl of cells were mixed with 5 μl of sample buffer, boiled for 5 min, and resolved by SDS/PAGE (25). Resolved proteins were transferred to nitrocellulose membrane (Bio-Rad) by transverse electrophoresis for 1 h at 500 mA in buffer containing 25 mM Tris·HCl (pH 8.3)/192 mM glycine/20% (vol/vol) methanol (26). Membranes were blocked overnight at 4°C in PBS with 3% BSA/0.05% Tween 20 and incubated with neutravidin coupled to horseradish peroxidase (Pierce) diluted 1:50,000 in the same buffer for 1 h at room temperature. The blots were washed extensively with PBS containing 0.02% Tween 20, developed by using the chemiluminescent substrate LumiGlo (Kirkegaard & Perry Laboratories), and exposed to x-ray film (Kodak XAR) for various times. Films were scanned by densitometry. The pixel density of the BtuB band was corrected for an equal background area and normalized to the labeling of the D6C protein, which was included on each gel as a labeling control. Multiple independent samples and exposure times were used to improve film response. This detection method is not quantitative but allows a consistent measure of the relative extent of labeling.
Immunofluorescence Microscopy. E. coli RK5016 cells harboring plasmids pBR322, pAG1btuB+, or pAG1 btuBV10P were grown as described above. To growing cells, E3R was added to 5 μg/ml and incubated for 15 min at 37°C. The bacteria were washed by centrifugation three times with PBS and suspended in 1 ml of PBS/3% BSA containing monoclonal mouse anti-HIS (Qiagen) (1:1,000). After 30 min at room temperature, cells were washed three times in PBS and suspended in 500 μl of PBS/3% BSA containing FITC-conjugated anti-mouse IgG antibody (1:500). After 30 min at room temperature, cells were washed three times in PBS and suspended in 100 μl of PBS. Samples were photographed in a Leica Leitz DMRBE fluorescence microscope by using a Photometrics SenSys camera.
Results
CNCbl Increases Reactivity of the N-Terminal Region of BtuB with Maleimide. The sulfhydryl-reactive reagent, BMCC (Pierce Endogen) was used to probe reaction with Cys substitutions at positions in the N-terminal region of BtuB (residues 1–17 and 25–31) and to detect changes in labeling in the presence of BtuB transport substrates. BMCC contains a maleimide moiety joined to biotin through a cyclohexyl-butyl linker. It can pass through OM porins but not through the cytoplasmic membrane. Covalent attachment of BMCC was detected by probing electrophoretically resolved cell proteins with horseradish peroxidase-coupled neutravidin. The degree of labeling at each position was compared with that of Cys at position 6 (D6C).
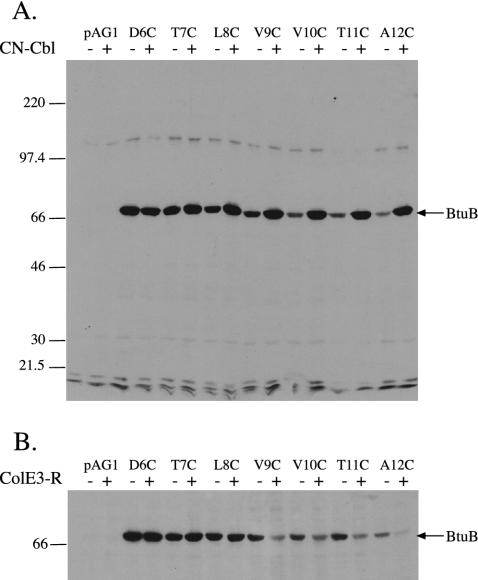
The addition of BMCC to intact cells resulted in almost exclusive labeling of BtuB proteins containing single Cys residues (Fig. 1A). Much lower signals resulted from labeling of a few other polypeptides or from normally biotinylated enzymes. Wild-type BtuB lacks Cys residues and was not labeled. Cys residues at positions 1–6 were labeled equally well in the absence or presence of CNCbl (Fig. 1 A and data not shown; labeling relative to D6C in Fig. 2A). Cys at positions 1–6 were maximally labeled, indicating that they are fully accessible to the periplasm, in agreement with the disordered state of residues 1–5 and high accessibility of residues 6 and 7 in the crystal structure (8). In the absence of CNCbl, positions 7–13 showed a progressive decrease in labeling by BMCC, down to 17% of the D6C level for N13C. The decreased labeling by BMCC of Cys residues beyond position 8 agrees with the crystal structure, which indicated that residues 9, 11, 12, and 16 have low accessibility. In contrast, the presence of CNCbl resulted in high labeling of these positions, comparable with the D6C level.
Fig. 1.
Reaction of BMCC with E. coli cells expressing BtuB with Cys substitutions. Strain RK5016 carrying pAG1 plasmid derivatives encoding BtuB variants with the indicated single Cys substitutions between positions 6 and 12 or the wild-type Cys-free protein were exposed to the indicated addition and BMCC (15 μg/ml) as described in Experimental Procedures. Cell extracts were resolved by SDS/PAGE, transferred to nitrocellulose membrane, probed with horseradish peroxidase-labeled neutravidin, and visualized by chemiluminescence. Molecular mass markers are indicated on the left in kilodaltons, and the position of BtuB is indicated on the right. (A) Addition of 5 μM CNCbl (lanes marked +). (B) Addition of E3R (+).
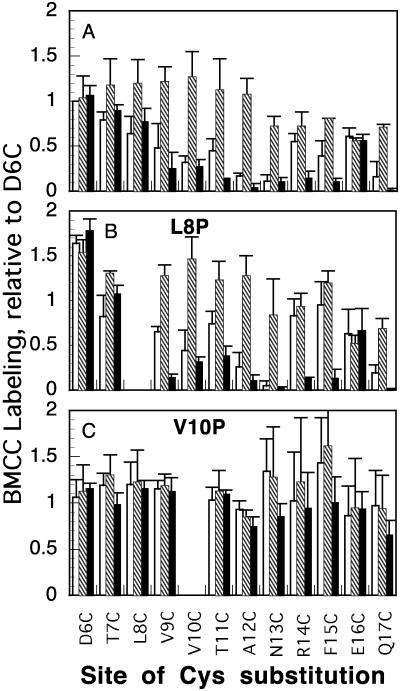
Fig. 2.
Effect of CNCbl and E3R on labeling of BtuB positions 6–17 by BMCC. Assays of BMCC labeling as described for Fig. 1 were subjected to densitometry, and the extent of label in the BtuB position was corrected for background density and normalized to the value for the D6C variant in the absence of substrates. Values are the average of at least six measurements taken from duplicate experiments with different exposure times; errors bar indicate standard error. Cells were incubated with no addition (open bars), CNCbl (hatched bars), and E3R (filled bars). The position of the Cys substitution is indicated on the bottom. The Cys substitutions were combined with no other substitutions (A), L8P (B), and V10P (C).
Labeling in the presence of CNCbl of positions 13–17 beyond the Ton box was decreased ≈30% relative to residues 1–12, suggesting impaired access to residues further in the barrel. In the absence of CNCbl, positions 13 and 17 were poorly labeled (≈15% of D6C), reflecting the conformational change after CNCbl binding seen in the Ton box. Positions 14–16 were almost equally accessible (≈50% of D6C) (Fig. 2 A) in the presence or absence of CNCbl. Thus, the CNCbl-induced conformational transition alters residues beyond the Ton box. In no case did CNCbl elicit decreased labeling than the control.
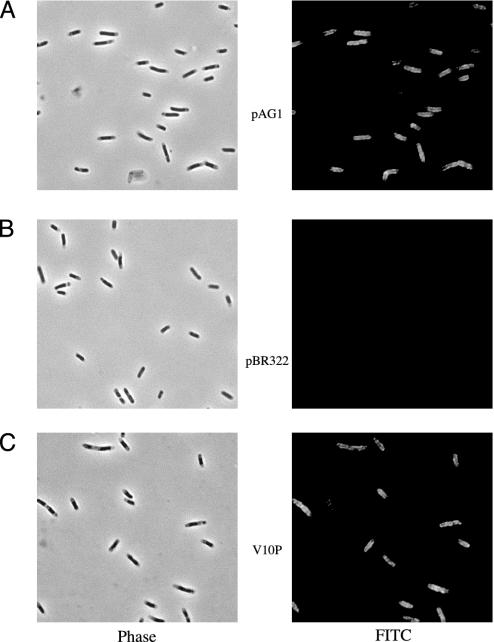
E3R Decreases Labeling of the Ton Box. We compared the effect of CNCbl on labeling with that of another BtuB substrate. A 76-aa receptor-binding R domain of colicin E9 binds BtuB with high affinity (12). The E3R has the same sequence and was expressed with a His6 tag to facilitate purification and detection. Binding of purified E3R to E. coli cells protects susceptible cells against challenge by colicin E3 (data not shown) and was visualized by immunofluorescence microscopy directed to the His6 tag (Fig. 3). Every cell carrying the btuB plasmid pAG1 bound E3R, and no cell-bound fluorescence was seen in the absence of E3R or in cells carrying the plasmid vector (Fig. 3B).
Fig. 3.
Immunofluorescence detection of binding of E3R to cells of E. coli expressing wild-type BtuB (pAG1) (A), the vector pBR322 (B), and the V10P variant of BtuB (C). Cells were exposed to the His6-tagged E3R domain, washed extensively, and then visualized with antisera to the His6 epitope followed by FITC-labeled secondary antibody. Phase contrast images (Left) and the FITC fluorescence (Right) of the same field are shown. There was no fluorescence in the absence of E3R.
The effect of E3R on reaction of BMCC with the Cys variants was quite different than that of CNCbl. In no case did E3R elicit higher labeling than in the control. As seen with CNCbl, positions 1–8 (data not shown and Figs. 1B and 2 A) were highly labeled and unaffected by E3R. Labeling of positions 9–12 (Figs. 1B and 2 A) was substantially decreased by E3R. Because E3R did not affect labeling of positions 1–8, it is unlikely that its inhibition of labeling at other positions was nonspecific. The decrease in labeling in response to E3R affected residues 13–17 except for position 16. Position 16 showed similar high labeling under all three conditions, consistent with EPR results (27) and its high accessibility in the crystal structure (8). Thus, E3R stabilizes a restricted or docked conformation in which the Ton box and distal residues are poorly accessible to labeling. This behavior matches the increased proportion of the immobile conformation seen in EPR analysis (28).
Effect of Uncoupling Mutations on Ton Box Labeling. The BtuB L8P and V10P substitutions confer the TonB-uncoupled phenotype (13, 14). Both uncoupling mutations affected the dynamics of the Ton box and its interaction with TonB. With both substitutions spin probes on the remaining Ton box positions exhibited high mobility independent of CNCbl (29). The uncoupling substitutions extended the range of Ton box positions able to form disulfide bonds to TonB (13). Increased formation of BtuB-TonB heterodimers in the presence of CNCbl occurred similar to the wild-type case with the L8P substitution but not with the V10P substitution. The uncoupling mutations might affect the docking of the Ton box to its restricted conformation differently.
The two uncoupling mutations had different effects on BMCC labeling. In the context of the L8P substitution, labeling of most positions between 6 and 15 was similar or higher than in the wild-type context (Fig. 2B) but showed the progressive decrease at the distal positions as in the wild-type case. The L8P Ton box showed increased labeling with CNCbl and decreased labeling with E3R (Fig. 2B). Possible distortion by the L8P substitution can be inferred from the increased labeling of many positions in the unliganded state. In contrast, the V10P substitution rendered all residues from 1 to 17 about equally accessible, and their high labeling was unaffected by either substrate (Fig. 2C). This constitutive labeling behavior is not the result of a defect in substrate binding, because the V10P mutant still bound CNCbl (14) and E3R (Fig. 3C). Thus, the L8P substitution allows substrate-induced transitions to occur, but V10P prevents formation of the docked conformation.
The TonB-uncoupled phenotype results from substitution of Pro or Gly at certain Ton box positions (13, 14), which could interfere with backbone conformation or side-chain recognition. To test whether Pro substitutions elsewhere in the Ton box conferred similar behavior, the D6P or A12P substitutions were combined with the T7C or V9C substitutions. All four double mutants exhibited normal BtuB transport activities (data not shown). The extent of labeling of either Cys residue by BMCC and the response to CNCbl or E3R were unaffected by the presence of either Pro substitution (data not shown). Thus, the ability of Pro substitutions in the Ton box to affect signal transduction and function is specific for their position.
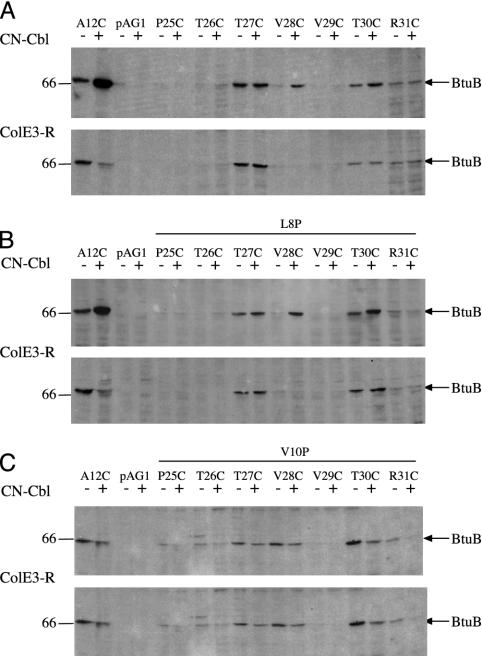
Labeling of Residues in Hatch Domain. Because substrates affected accessibility of residues beyond the Ton box to residue 17, we examined the labeling of positions 25–31. This region resembles the Ton box in sequence (TTVVTR compared with TLVVTA of the Ton box) but is the first strand of the β-sheet that forms the central core of the hatch domain (8). Residues in this strand were much less reactive with BMCC than were the upstream residues, reflecting its location further in the barrel. Positions 25, 26, and 29 were scarcely labeled even in the presence of substrates or the L8P substitution (Fig. 4). The V10P substitution conferred a slight increase in labeling. In contrast, positions 27 and 30 were weakly labeled under all conditions. Position 30 was somewhat more accessible in the presence of CNCbl but did not respond to E3R. Position 28 behaved similar to Ton box residue 12. Position 31 was unusual in that its labeling was lower in the presence of CNCbl in the uncoupled mutants. Thus, the transitions of the Ton box are reflected in changes of access of positions in the core structure of the hatch, but most of these residues are poorly accessible or reactive and undergo no obvious substrate-induced conformational changes (8).
Fig. 4.
Reaction of BMCC with BtuB carrying Cys substitutions in positions 25–31. Included controls as indicated were A12C and wild-type BtuB in pAG1. The Cys substitutions were combined with no other substitution (A), L8P (B), and V10P (C). (A–C Upper) Effect of addition of CNCbl. (A–C Lower) Effect of E3R.
Finally, to test whether the changes in labeling are responses to substrate binding or transport, BMCC labeling was carried out as described above with the panel of Cys variants expressed in a tonB host strain incapable of CNCbl uptake (data not shown). There was no detectable difference in labeling of any of the positions or their response to CNCbl or E3R related to TonB function.
Discussion
The TonB system is an unusual process of active transport present only in Gram-negative bacteria. It provides energy to transporters in the OM which cannot maintain ion gradients and is physically separate from ATP pools. How TonB acts is still a matter of conjecture. Its N-terminal region associates with the ExbBD complex in the cytoplasmic membrane, and its C-terminal domain associates with the OM. Although TonB contacts both membranes, there is evidence that it may shuttle between them and release contact with the cytoplasmic membrane to bind to OM targets (30, 31).
TonB action involves its recognition of the Ton box on the transporters, probably in response to a conformational transition that follows substrate binding. TonB may also interact with other parts of the transporters, as implied from genetic suppression patterns (15). Claims that FepA and FhuA variants lacking the hatch domain and Ton box (32, 33) still exhibit TonB-dependent activity have been challenged by the recognition that the empty barrels can be complemented by the hatch domains encoded by the mutated chromosomal alleles (34). The N-terminal regions of FhuA and FecA move extensively after substrate binding, but the Ton box regions themselves are disordered. Because of the absence of a switch helix in FepA and BtuB, the behavior of their Ton boxes cannot be inferred from the behavior of FepA and FecA. Only in the BtuB structures can the Ton box be seen in the presence and absence of CNCbl (8). Unexpectedly, the Ton boxes of the empty and substrate-loaded forms had similar conformations and no indication of extension into the periplasm. The crystallographic views were thus inconsistent with results obtained by disulfide trapping and site-directed spin labeling (13, 17, 23, 29), which indicated that CNCbl triggers a substantial increase in residue mobility and contact of Ton box residues with TonB.
Two major conclusions can be drawn from the BMCC labeling. The more surprising finding was that two BtuB substrates elicit different conformations of the Ton box. CNCbl conferred increased access to labeling at many positions, whereas E3R resulted in decreased access. Site-directed spin labeling (28) revealed the same behavior in that CNCbl and E3R conferred increased and decreased mobility of the residues, respectively. The EPR spectra were consistent with the presence of two conformations of markedly different residue mobility, the fractional distribution of which was shifted by the transport substrates. Comparison of our results equates the low-mobility state with low labeling by BMCC and the high-mobility state with higher labeling. The two-state model requires that in the absence of ligands, the Ton box fluctuates between both states such that BMCC labeling occurs during transient appearance of the mobile state.
The second conclusion helps reconcile previous findings. Labeling showed that the N-terminal residues 1–6 were highly accessible and little affected by substrate binding, in agreement with their high mobility in EPR (29) and their disorder in the crystal structure (8). The crystal structures show that the Ton box does not form a regular secondary structure but is nestled between other structural elements (8). The progressive decrease in labeling of residues 7–15 is consistent with the relative accessibility of these residues in the crystal structure, calculated by David Chimento (University of Virginia) (8). Calculated access to probes of different sizes (1.4- or 2.1-Å diameter) showed no differences except for positions 13–15 and 24, which were inaccessible to the larger probe. Major differences between the unbound and CNCbl-bound form were seen only at residues 6, 7, 20, and 26. Residue access was related to the degree of BMCC labeling, except labeling of positions 9, 11, 14, and 15 was higher than expected for accessibility. This difference may be related to dynamic opening of those residues. Thus, BMCC labeling agrees reasonably well with the crystal structure of the unbound form of BtuB. The high labeling in the presence of CNCbl is inconsistent with the crystal structure but agrees well with the EPR results. Perhaps the extension of the Ton box is affected by the crystallization conditions.
Although EPR results with holo-BtuB agree with BMCC labeling, there are considerable differences with apo-BtuB, in which sharply alternating patterns of residue mobility are revealed by EPR (23, 29). An explanation for this behavior is that attachment of the spin probe at positions of low solvent accessibility, namely residues 9, 11, and 12, interferes with formation of the immobile, docked state and hence locks the region into its mobile state. Substitution of Thr-11 to Phe to mimic the spin probe resulted in conversion of position 7 to the high-mobility state (27). EPR studies showed that switching between the mobile and immobile forms occurs with little barrier and can be triggered by changes in the bilayer environment, certain mutations, and substrate binding (35). Taken together, these findings now support a consistent scenario. In the apo form of BtuB, the Ton box prefers to dock into a conformation that exhibits low backbone mobility and low accessibility to labeling by BMCC. Binding of E3R stabilizes the docked conformation and binding of CNCbl results in undocking, which is characterized by high residue mobility and high labeling. Hence, E3R must make different contracts with BtuB than does CNCbl, although some contact sites are likely shared. The finding the E3R confers a less accessible configuration suggests that the colicin probably does not cross the OM through the BtuB barrel.
Properties of the L8P and V10P uncoupling mutations can be addressed. The L8P substitution had little effect on substrate-induced transitions of the Ton box detected by BMCC labeling, although it resulted in high mobility of the Ton box measured by EPR. Perhaps the presence of both a spin probe and the uncoupling mutation destabilizes the V8P Ton box to result in the undocked conformation. In contrast, the V10P substitution confers high accessibility and mobility with no response to substrate addition in both assays. Thus, the V10P variant is unable to form the docked immobile state under any condition tried. Presumably, the defect in the L8P substitution does not affect the substrate-induced transitions but interaction with TonB. In summary, the value of combining multiple approaches to test signal transduction is apparent.
Acknowledgments
Insights into BtuB structure by David Chimento, Michael Wiener, and Gail Fanucci are gratefully acknowledged. This work was supported by National Institutes of Health Research Grants GM19078 (to R.J.K.) and GM35215 (to D.S.C.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: OM, outer membrane; Cbl, cobalamin; CNCbl, cyano-Cbl; BMCC, 1-biotinamido-4-[4′-(maleimidomethyl)cyclohexane-carboxamido]butane; E3R, receptor-binding domain of colicin E3.
References
- 1.Braun, V. & Braun, M. (2002) Curr. Opin. Microbiol. 5, 194–201. [DOI] [PubMed] [Google Scholar]
- 2.Faraldo-Gomez, J. D. & Sansom, M. S. P. (2003) Nat. Rev. Mol. Cell Biol. 4, 105–116. [DOI] [PubMed] [Google Scholar]
- 3.Ferguson, A. D. & Deisenhofer, J. (2002) Biochim. Biophys. Acta 1565, 318–332. [DOI] [PubMed] [Google Scholar]
- 4.Ferguson, A. D., Hofmann, E., Coulton, J. W., Diederichs, K. & Welte, W. (1998) Science 282, 2215–2220. [DOI] [PubMed] [Google Scholar]
- 5.Locher, K. P., Rees, B., Koebnik, R., Mitschler, A., Moulinier, L., Rosenbusch, J. P. & Moras, D. (1998) Cell 95, 771–778. [DOI] [PubMed] [Google Scholar]
- 6.Buchanan, S. K., Smith, B. S., Venkatramani, L., Xia, D., Esser, L., Palnitkar, M., Chakraborty, R., van der Helm, D. & Deisenhofer, J. (1999) Nat. Struct. Biol. 6, 56–63. [DOI] [PubMed] [Google Scholar]
- 7.Ferguson, A. D., Chakraborty, R., Smith, B. S., Esser, L., van der Helm, D. & Deisenhofer, J. (2002) Science 295, 1715–1719. [DOI] [PubMed] [Google Scholar]
- 8.Chimento, D. P., Mohanty, A. K., Kadner, R. J. & Wiener, M. C. (2003) Nat. Struct. Biol. 10, 394–401. [DOI] [PubMed] [Google Scholar]
- 9.James, R., Kleanthous, C. & Moore, G. R. (1996) Microbiology 142, 1569–1580. [DOI] [PubMed] [Google Scholar]
- 10.Lazdunski, C. J., Bouveret, E., Rigal, A., Journet, L., Lloubes, R. & Benedetti, H. (1998) J. Bacteriol. 180, 4993–5002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DiMasi, D. R., White, J. S., Schnaitman, C. A. & Bradbeer, C. (1973) J. Bacteriol. 115, 506–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Penfold, C. N., Garinot-Schneider, C., Hemmings, A. M., Moore, G. R., Kleanthous, C. & James, R. (2000) Mol. Microbiol. 38, 639–649. [DOI] [PubMed] [Google Scholar]
- 13.Cadieux, N., Bradbeer, C. & Kadner, R. J. (2001) J. Bacteriol. 182, 5954–5961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gudmundsdottir, A., Bell, P. E., Lundrigan, M. D., Bradbeer, C. & Kadner, R. J. (1989) J. Bacteriol. 171, 6526–6533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bell, P. E., Nau, C. D., Brown, J. T., Konisky, J. & Kadner, R. J. (1990) J. Bacteriol. 172, 3826–3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heller, K. J., Kadner, R. J. & Gunther, K. (1988) Gene 64, 147–153. [DOI] [PubMed] [Google Scholar]
- 17.Cadieux, N. & Kadner, R. J. (1999) Proc. Natl. Acad. Sci. USA 96, 10673–10678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ogierman, M. & Braun, V. (2003) J. Bacteriol. 185, 1870–1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moeck, G. S., Tawa, P., Xiang, H., Ismail, A. A., Turnbull, J. L. & Coulton, J. W. (1996) Mol. Microbiol. 22, 459–471. [DOI] [PubMed] [Google Scholar]
- 20.Moeck, G. S., Coulton, J. W. & Postle, K. (1997) J. Biol. Chem. 272, 28391–28397. [DOI] [PubMed] [Google Scholar]
- 21.Moeck, G. S. & Letellier, L. (2001) J. Bacteriol. 183, 2755–2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hubbell, W. L., Cafiso, D. S. & Alenbach, C. (2000) Nat. Struct. Biol. 7, 735–739. [DOI] [PubMed] [Google Scholar]
- 23.Merianos, H. J., Cadieux, N., Lin, C. H., Kadner, R. J. & Cafiso, D. S. (2000) Nat. Struct. Biol. 7, 205–209. [DOI] [PubMed] [Google Scholar]
- 24.Reynolds, P. R., Mottur, G. P. & Bradbeer, C. (1980) J. Biol. Chem. 255, 4313–4319. [PubMed] [Google Scholar]
- 25.Laemmli, U. K. (1970) Nature 227, 680–685. [DOI] [PubMed] [Google Scholar]
- 26.Towbin, H., Staehlin, T. & Gordon, J. (1979) Proc. Natl. Acad. Sci. USA 76, 4350–4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fanucci, G. E., Coggshall, K. A., Cadieux, N., Kim, M., Kadner, R. J. & Cafiso, D. S. (2003) Biochemistry 42, 1391–1400. [DOI] [PubMed] [Google Scholar]
- 28.Fanucci, G. E., Cadieux, N., Kadner, R. J. & Cafiso, D. S. (2003) Proc. Natl. Acad. Sci. USA, in press. [DOI] [PMC free article] [PubMed]
- 29.Coggshall, K. A., Cadieux, N., Piedmont, C., Kadner, R. J. & Cafiso, D. S. (2001) Biochemistry 40, 13964–13971. [DOI] [PubMed] [Google Scholar]
- 30.Letain, T. E. & Postle, K. (1997) Mol. Microbiol. 24, 271–283. [DOI] [PubMed] [Google Scholar]
- 31.Larsen, R. A., Letain, T. F. & Postle, K. (2003) Mol. Microbiol. 49, 211–218. [DOI] [PubMed] [Google Scholar]
- 32.Braun, M., Killmann, H. & Braun, V. (1999) Mol. Microbiol. 33, 1037–1049. [DOI] [PubMed] [Google Scholar]
- 33.Scott, D. C., Cao, Z., Qi, Z., Bauler, M., Igo, J. D., Newton, S. M. & Klebba, P. E. (2001) J. Biol. Chem. 276, 13025–13033. [DOI] [PubMed] [Google Scholar]
- 34.Vakharia, H. & Postle, K. (2002) J. Bacteriol. 184, 5508–5512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fanucci, G. E., Cadieux, N., Piedmont, C. A., Kadner, R. J. & Cafiso, D. S. (2002) Biochemistry 41, 11543–11551. [DOI] [PubMed] [Google Scholar]