Abstract
Ionizing radiation (IR) is a known human breast carcinogen. Although the mutagenic capacity of IR is widely acknowledged as the basis for its action as a carcinogen, we and others have shown that IR can also induce growth factors and extracellular matrix remodeling. As a consequence, we have proposed that an additional factor contributing to IR carcinogenesis is the potential disruption of critical constraints that are imposed by normal cell interactions. To test this hypothesis, we asked whether IR affected the ability of nonmalignant human mammary epithelial cells (HMEC) to undergo tissue-specific morphogenesis in culture by using confocal microscopy and imaging bioinformatics. We found that irradiated single HMEC gave rise to colonies exhibiting decreased localization of E-cadherin, β-catenin, and connexin-43, proteins necessary for the establishment of polarity and communication. Severely compromised acinar organization was manifested by the majority of irradiated HMEC progeny as quantified by image analysis. Disrupted cell–cell communication, aberrant cell–extracellular matrix interactions, and loss of tissue-specific architecture observed in the daughters of irradiated HMEC are characteristic of neoplastic progression. These data point to a heritable, nonmutational mechanism whereby IR compromises cell polarity and multicellular organization.
Epidemiologic data indicate that women exposed to ionizing radiation (IR) for either therapy (1, 2), diagnostic purposes (3), or as a consequence of atomic bombs (4) have an increased risk of breast cancer. The action of IR as a DNA-damaging agent, and consequently as a mutagen, is widely considered to be the basis for its action as a carcinogen (5). However, tissue response to radiation, and hence risk, is a composite of genetic damage and epigenetic events, such as altered intercellular communicaton (6). Recent experimental models suggest that carcinogenesis can be driven by abnormal interactions between cells and their microenvironment (reviewed in refs. 7 and 8). We have shown that irradiated mammary stroma promotes tumorigenesis of unirradiated mammary epithelial cells (9), and that transforming growth factor β1 (TGF-β) activation mediates cellular and tissue response to IR (10–12). Thus, in addition to causing DNA damage, radiation exposure alters key regulators of cell phenotype that affect, directly or indirectly, the ability of normal tissue to suppress abnormal cell growth (13).
Epithelial cells depend on signals from the microenvironment to establish the requisite polarity for functional differentiation (14). Release from these constraints has profound consequences on tumorigenesis, progression, and metastasis (reviewed in refs. 6, 8, and 15). Tumorigenic and nontumorigenic human mammary epithelial cells (HMEC) are nearly indistinguishable when cultured as monolayers, but readily diverge in terms of morphogenesis in an appropriate microenvironment, which is evident in a three-dimensional reconstituted basement membrane (3D rBM) assay that we developed (16). In this assay, nonmalignant HMEC arrest growth and form lumen-containing acini similar to those found in situ, whereas breast cancer cells continue to proliferate and aggregate, rather than organize. Formation of acini requires expression and appropriate localization of proteins involved in the establishment of tissue structure and polarity (17). HMEC colonies that develop into phenotypically normal acini exhibit among other markers, E-cadherin at the interface between cells, basolateral β1-integrin, and basal α6-integrin (18). In contrast, breast cancer colonies exhibit disorganized, decreased, or aberrant expression of these markers, similar to what is observed in primary breast cancer.
If radiation exposure affects not only the phenotype of stromal but also of epithelial cells, such alterations could potentially promote neoplastic progression in susceptible cells. To test this hypothesis, we asked whether sublethal IR doses perturbed the ability of HMEC to undergo mammary-specific morphogenesis in a physiological context by using the 3D rBM assay. To replicate a key component of the irradiated stroma (10–12), TGF-β was added to some cultures. To measure the global consequences of irradiation, we used confocal microscopy and an imaging bioinformatics system for integrated image acquisition, annotation, and hierarchical image abstraction to register localization and expression information of targets along with positional references and morphological features (19).
We found that irradiated single HMEC gave rise to colonies where nearly all progeny failed to establish basal polarity and lost organizational integrity as measured by several parameters. As shown by quantitative image analysis, these changes were shared by the majority of the population. This finding is inconsistent with a radiation-induced mutational mechanism, which was confirmed by the absence of measurable changes in the population genome analyzed by comparative genomic hybridization analysis. Moreover, because the phenotype is exhibited by the daughters of individually irradiated cells, these data suggest that radiation causes a heritable alteration in pathways affecting cell adhesion, extracellular matrix (ECM) interactions, epithelial polarity, and cell–cell communication. Thus, epigenetic events after radiation exposure disrupt multicellular organization, which we postulate will override the positive influence of tissue architecture that usually impedes neoplastic progression.
Methods
Cell Culture. HMT-3522-S1 human mammary epithelial cells (S1; passages 53–60) were grown as described (18). Although phenotypically normal and nonmalignant, the S1 are an established cell line that have a number of chromosomal changes and an extended life span in culture (20). S1 cell monolayers were grown until 70% confluent before trypsinization, and single cells (8 × 105 cells per ml) were embedded into Matrigel (Collaborative Research) with or without 400 pg/ml recombinant human TGFβ1 (R & D Systems) and irradiated within 5 h by using 60Co γ-radiation at a dose rate of 90 cGy/min to a total dose of 2 Gy. Dosimetry was determined by using a Victoreen ionization chamber. Control plates were sham irradiated. Media were changed on alternate days. Cells were grown in the presence of epidermal growth factor for 6 days, and harvested at 10 days. For immunocytochemistry cultures were embedded in Tissue-Tek compound (Sakura Finetek, Torrance, CA), and frozen in a dry ice/ethanol bath. Blocks were stored frozen until time of sectioning.
Immunofluorescence. Cryosections (20 μm) were cut at –30°C onto gelatin-coated coverslips. Sections were fixed by using methanol/acetone (1:3) at –20°C for 10 min or 4% paraformaldehyde for E-cadherin. Nonspecific sites were blocked by using the supernatant from a 0.5% casein/PBS (pH 7.4) solution for 1 h at room temperature (RT). Sections were incubated in primary antibody diluted in blocking buffer for 1 h at RT in a humidified chamber. Antibodies used were rat anti-mouse CD29 (Pharmingen) to integrin β1 chain monoclonal antibody, rat anti-human CD49f monoclonal antibody (Pharmingen) to integrin α6 chain, and mouse monoclonal antibody to E-cadherin (BD Transduction Laboratories). Sections were washed in PBS containing 0.1% BSA, before incubating in secondary antibody conjugated to Alexa Fluor 488 (Molecular Probes) for 1 h at RT in a dark humidified chamber, washed, and counterstained with TO-PRO-3 iodide (Molecular Probes), before mounting with Vectasheild mounting medium (Vector Laboratories, Burlingame, CA).
Image Acquisition, Processing, and Analysis. Dual immunofluorescence confocal images were acquired by using a Zeiss LSM 410 inverted laser scanning confocal microscope equipped with an external argon/krypton laser. Confocal images were captured at 0.5-μm intervals as 8-bit images by using a Zeiss Fluor ×40 (1.3 numerical aperture) objective. Images were standardized by comparing only images stained with the same antibodies in the same experiment, captured with the same parameters at the same times, and scaled and displayed identically. Relative intensity of images was scaled by using scilimage (TNO Institute of Applied Physics, Delft, The Netherlands), which was used to define a standard sized region of the TO-PRO-3 iodide image (nuclei slice) without reference to the Alexa Fluor 488 images. Statistical significance of the mean fluorescence intensity for each region of interest (n = 20 colonies) and standard error for each treatment group was determined by using the unpaired Student's t test (prism, GraphPad, San Diego). The displayed images were those closest to the mean intensity for the treatment group.
Segmentation of nuclei was used to determine acinar organization at the colony midsection (21). This model-based approach assumes that the projection of each nucleus is quadratic in the image space. Instead of grouping step and roof edges, the segmentation is initiated from a representation that corresponds to the zero crossings of the image. The zero crossing image is then filtered with geometrical and illumination constraints to form binarized clump of nuclei, which is then partitioned into several nuclei through a process that is called centroid transform.
Protein Extraction and Immunoblotting. Cells in the 3D rBM assay were isolated by ice-cold PBS/EDTA (0.01 M sodium phosphate, pH 7.2, containing 138 mM sodium chloride and 5 mM EDTA) (18) and lysed in buffer as described (18). Equal amounts of protein lysates were run on reducing SDS/PAGE and then immunoblotted and detected by using a Pierce Super-Signal system (Pierce). Blots were also probed for β-actin to assess equal loading of protein. Exposed films were scanned and subjected to densitometric analysis for the determination of relative amount.
Comparative Genomic Hybridization. Array comparative genomic hybridization was performed at the University of California, San Francisco, Cancer Center as described (22). Briefly, 1 μg each of test and reference genomic DNAs were fragmented by DPNII digestion, labeled by random priming with CY3- and CY5-dUTP (Amersham Pharmacia), respectively, coprecipitated with 80 μg of human cot-1 DNA (Life Technologies), and resuspended in 20 μl of hybridization buffer (50% formamide/10% dextran sulfate/2× SSC/2% SDS/200 g of yeast tRNA). This mixture was denatured at 75°C for 10 min followed by 60 min at 37°C. Just before hybridization, array slides were processed following the manufacturer's recommendations (Surmodics, Eden Prairie, MN). A frameseal frame was placed around each array, hybridization mix was added, and the slide was placed in a plastic slide holder, prewarmed to 37°C, containing 200 μl of wash buffer (50% formamide/2× SSC) to prevent evaporation. Hybridization was carried out at 37°C for 48–72 h on a gently rocking platform. After hybridization, slides were immersed for 15 min at 48°C in wash buffer, followed by washes at 48°C in 2× SSC, 0.1% SDS for 30 min, and 0.1 M sodium phosphate buffer containing 0.1% Nonidet P-40, pH 8.0, at RT for 10 min. Slides were then rinsed in 2× SSC and dried by centrifugation.
Results
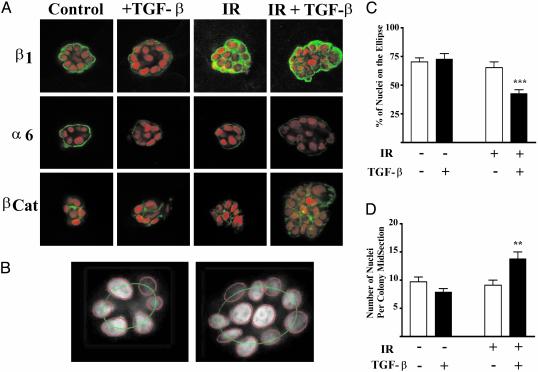
Progeny of Irradiated Cells Exhibit Perturbed Cell–ECM and Cell–Cell Adhesion. To determine whether IR alters the ability of epithelial cells to functionally interact with the microenvironment, we used the 3D rBM assay of morphogenesis in a laminin-rich basement membrane where changes in tissue structure can be quantified (16). Single HMT-3522 S1 human mammary epithelial cells were cultured, with and without the addition of TGF-β, and irradiated with a dose of 2 Gy, except where noted, 3–5 h after plating in Matrigel. Surviving cells, ≈80% (not shown), formed multicellular colonies over 5–7 days and then underwent morphogenesis into hollow spheres that recapitulate mammary acini by day 10. Immunofluorescence of β1 and α6 integrins and β-catenin at the colony mid-section were analyzed by using confocal microscopy (Fig. 1A). HMEC colonies express basolateral β1-integrin and basal α6-integrin, which are critical for acinar organization (23). HMEC colonies arising from irradiated cells exhibited increased β1-integrin immunoreactivity that was distributed throughout the cytoplasm (Fig. 1 A). In contrast, the immunoreactivity of α6-integrin, which partners with β4 integrin, was decreased in colonies generated from irradiated cells. A collagen IV-containing basement membrane was observed in all treatment groups, indicating that changes in integrin expression were not caused by the lack of appropriate ligand for this ECM receptor (not shown). Treatment with TGF-β did not alter β1 integrin localization but did reduce α6 integrin immunoreactivity further. β-catenin, which is involved in cell–cell adhesion via the cytoskeleton and E-cadherin, was localized to the lateral cell borders in colonies from nonirradiated cells. β-catenin immunoreactivity was decreased in colonies derived from irradiated cells.
Fig. 1.
Perturbed protein localization and acinar organization as a function of TGF-β, IR, and dual treatment. Colonies develop and organize in 3D rBM culture over the course of 10 days, during which time the cells are fed every other day. EGF, to stimulate proliferation, is removed at day 6, and the cells are harvested at day 10. (A) Representative images of colonies from control, TGF-β-, IR-, or dual-treated cultures. The image is representative of the mean intensity for each marker based on image analysis of 20 randomly chosen colonies. Immunostaining of β1 integrin, α6 integrin, and β-catenin was detected by using secondary antibodies labeled with Alexa Fluor 488 (green). Nuclei are counterstained with TO-PROR-3 iodide, shown in red. Note the loss of acinar organization in the irradiated colonies. (B) Acinar organization was measured by nuclear segmentation of the colony confocal midsection fit to an ellipse as shown for a control (Left) and dual-treated (Right) colony. (C) Acinar organization as a function of treatment group (n > 100 colonies per treatment). Acinar organization was significantly (P < 0.0001) decreased in colonies that arose from irradiated cells that were cultured in the presence of TGF-β.(D) The number of nuclei per colony midsection as a function of treatment group. The number of nuclei was significantly (P < 0.001) increased in colonies arising from irradiated cells treated with TGF-β.
Disrupted Tissue-Specific Morphogenesis and the Irradiated Phenotype as Quantified by Image Analysis Reveal a Global HMEC Response. The use of morphogenesis as a readout of cellular function requires systematic analysis of colony organization and protein localization to classify the degree of response. It is therefore desirable to conduct population studies and correlate features measured from images of cells with their treatment. The acinar-like organization of colonies was analyzed by using the relative nuclear position in confocal optical midsection as described in Methods (Fig. 1 B–D). The degree of acinar organization around a central lumen was determined by fitting the nuclei to an ellipse (Fig. 1B). Acinar organization was significantly (P < 0.0001) reduced in colonies arising from irradiated cells that were cultured with TGF-β. The number of cells per midsection was also significantly increased (P < 0.001) in irradiated, TGFβ-treated HMEC colonies in comparison to colonies from control cells or those exposed to single agents.
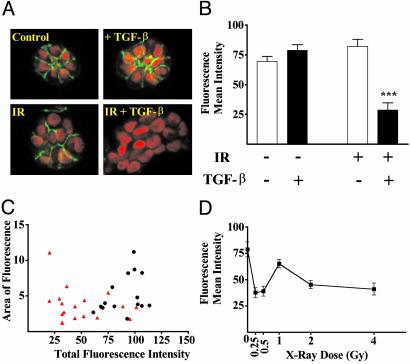
The assembly of cells into tissue-specific structures requires the interaction of different cell adhesion systems. E-cadherin is a crucial epithelial adhesion molecule that links cells via an homophilic extracellular domain and is anchored intracellularly to the cytoskeleton via dynamic interactions with the catenins (24). Low E-cadherin immunoreactivity in breast cancer is associated with poor prognosis (25), whereas restoration of E-cadherin reverts the invasive phenotype of cancer cells (26). We localized E-cadherin by using immunofluorescence, confocal microscopy and image analysis (Fig. 2). Colonies from irradiated cells cultured in the presence of TGFβ showed a significant (P < 0.0001) loss of E-cadherin immunoreactivity compared with control cells. The unlikely possibility that the colonies surviving treatment were selected from a previously existing population was addressed by examining the distribution of individual colonies within each treatment group in comparison to control colonies. A representative analysis is shown for E-cadherin, indicating that the dual treated colonies form a distinct population (Fig. 2C). To determine whether the effect on cell interactions was sensitive to radiation dose, we performed a dose–response (Fig. 2D). E-cadherin immunoreactivity was significantly decreased in colonies arising from cells exposed to as little as 25 cGy, a dose that does not result in appreciable cell kill. To determine whether radiation exposure and TGF-β treatment resulted in significant changes in the genomic sequence, we performed comparative genomic hybridization as described in Methods. This analysis did not reveal any significant differences between the untreated and double-treated populations (data not shown), which supports the global population response revealed by quantitative image analysis.
Fig. 2.
E-cadherin immunoreactivity and localization are significantly reduced by IR and TGF-β.(A) Confocal images of E-cadherin immunoreactivity in midsections of colonies representative of the average response as measured by image analysis of 20 colonies are shown for each treatment group. E-cadherin (green) and nuclei (red) were detected as described in Fig. 1. (B) Quantified E-cadherin immunoreactivity as a function of treatment group. The mean intensity of E-cadherin immunofluorescence was significantly (P < 0.0001) reduced in TGF-β-treated, irradiated colonies. (C) Display of relative intensity versus colony area for sham (black circles) and dual-treated (red triangles) colonies. Comparison of the treated to control populations show that >75% of the treated colonies exhibit loss of E-cadherin, a frequency that cannot be explained by mutation rates. (D) Quantified E-cadherin immunoreactivity as a function of radiation exposure. The dose–response shows significant loss of E-cadherin immunoreactivity at doses that do not lead to any detectable loss of cell viability.
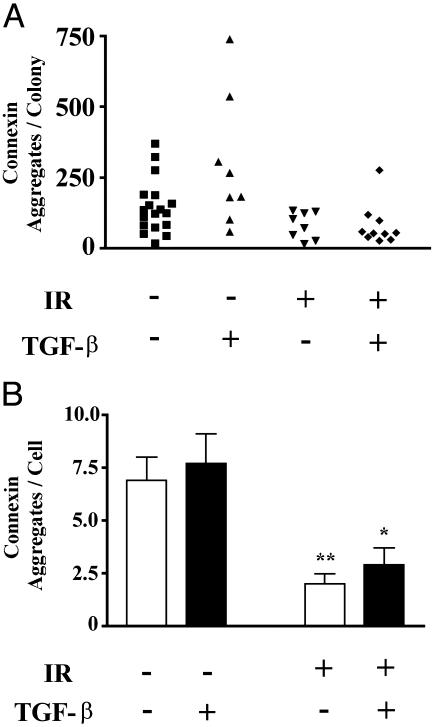
There is an intricate relationship between cell–ECM and cell–cell adhesion in glandular tissues. To determine whether other cell–cell adhesion molecules also change, we measured connexin 43, a member of a family of proteins that assemble into gap junctions and modulate the transfer of molecules between cells. Breakdown of gap junctional complexes is induced by tumor promoters (27) and correlate with breast cancer metastatic potential (28, 29). Connexin 43 is also associated with the function and signaling of E-cadherin (30, 31). In S1 HMEC acinar colonies, connexin-43 was localized as distinct aggregates between cells. The number of connexin 43 foci per colony decreased after radiation exposure, regardless of TGF-β exposure (Fig. 3). When normalized to the number of cells per colony, the frequency of connexin foci decreased >3-fold in the daughters of irradiated cells (2.0 ± 0.46, n = 8) compared with those from unirradiated cells (6.9 ± 1.1, n = 18).
Fig. 3.
Gap junctions are decreased in irradiated colonies. (A) Connexin 43 was localized by immunostaining and randomly selected colonies were imaged by confocal microscopy. Data shown are representative of two independent experiments. The number of aggregates per colony are displayed for 8–18 colonies per treatment. (B) The average (±SE) number of connexin 43 foci per cell is displayed as a function of treatment group. Colonies arising from irradiated cells showed significantly (P < 0.05, two-tailed t test) fewer connexin foci than those from nonirradiated cells.
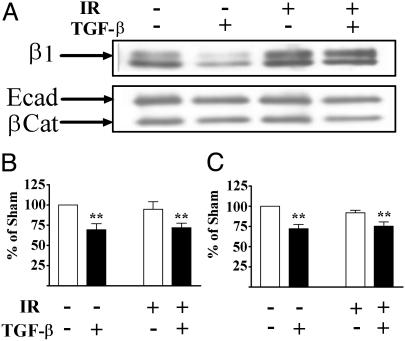
Decreased E-Cadherin and β-Catenin Localization Are Not a Function of Protein Abundance. E-cadherin localization can be modified by the degree of association with the cytoskeleton via the catenins. β-catenin and E-cadherin partner to link cells and the cytoskeleton via the adherens junction (32). To test whether decreased immunolocalization was caused by a change in the compartmentalization of these adhesion molecules, sections were detergent extracted to remove the soluble fraction. Detergent extraction before fixation did not alter the pattern or intensity of E-cadherin in dual-treated samples (data not shown). Consistent with this finding, immunoblotting total protein extracts showed that both E-cadherin and β-catenin levels were decreased in TGF-β-treated colonies (Fig. 4A). β1 integrin protein abundance, on the other hand, increased in irradiated samples regardless of TGF-β exposure, which is consistent with the increased cytoplasmic staining shown in Fig. 1B. In contrast, only the progeny of irradiated cells showed a decrease in E-cadherin immunolocalization. These data suggest that decreased E-cadherin immunoreactivity at the cell junctions in the dual treated colonies reflects both a TGF-β-induced decrease in protein levels and a radiation-induced change in localization, suggesting a change in complex formation at the cell surface.
Fig. 4.
Protein levels as a function of IR and TGF-β. (A) Representative immunoblots of β1 integrin, E-cadherin, and β-catenin from total protein extracted from cultures. β1 integrin protein abundance increased in irradiated samples, regardless of TGF-β exposure. E-cadherin and β-catenin protein abundance were decreased in extracts from TGF-β-treated cultures, regardless of irradiation. Quantitation of E-cadherin (B) and β-catenin (C) protein abundance from three independent experiments normalized to β-actin are shown as mean and standard error. The protein levels in cell extracts were significantly (P < 0.01) reduced in TGF-β-treated cultures.
Together, these data indicate that IR can generate a persistent phenotype in daughter cells characterized by increased cytoplasmic β1 integrin, decreased α6 integrin, radically decreased cell surface localization of E-cadherin and β-catenin, and loss of connexin 43. The cumulative epigenetic changes in phenotype results in a loss of tissue-specific architecture that is indicative of malignant progression.
Discussion
In this study we show that irradiated single HMEC gave rise to colonies that had more cells, failed to establish tissue-specific organization, and expressed significantly less E-cadherin, β-catenin, and connexin-43. It is remarkable that the phenotype was exhibited by progeny of individually irradiated cells, suggesting that IR causes heritable alterations in pathways affecting cell adhesion, ECM interactions, epithelial polarity, and cell–cell communication. Release from cell–cell interactions, as demonstrated by experimentally induced loss and restitution of E-cadherin (26, 33), has profound consequences for breast cancer tumorigenesis, progression, and metastasis. The features of individual colonies measured by quantitative image analysis showed that these changes were present in the majority of the population, a finding inconsistent with the frequency of radiation-induced mutations and confirmed by the absence of measurable changes in the population genome. Thus epigenetic mechanisms initiated by irradiation of HMEC result in a malignant-like phenotype in progeny generations after IR exposure.
Intercellular and extracellular signals are critical to the suppression of neoplastic cellular behavior. Disruption of cell–cell interactions are implicated, if not required, in neoplastic progression (7, 8, 34). Radiation exposure alters the expression of many genes involved in tissue processes such as proteases, growth factors, cytokines, and adhesion proteins, which supports the view that carcinogenesis could compromise tissue integrity by altering the flow of information among cells (35, 36). Indeed, our recent experimental studies demonstrate that multicellular architecture can be dominant over genomic change in terms of malignant cellular behavior (18, 37, 38). In these studies, breast cancer cells treated with β1 integrin function-blocking antibodies revert from disorganized colonies to organized acinar-like colonies that are characterized by restoration of cytoskeletal organization, cell–cell and cell–ECM interactions, and reduced tumorigenecity (18). Small molecule inhibitors can also be used to cooperatively block aberrant signaling and revert tumorigenic behavior (37, 38). These data, and others in hematopoetic cancers (39), suggest that cancer can be controlled by reestablishing appropriate contacts from the ECM and stroma via outside-in signaling.
Although radiation can acutely regulate E-cadherin and α-catenin levels (40), as well as integrin expression (41), in our studies the phenotype is exhibited by the daughters of irradiated cells several generations after radiation exposure. The redistribution of β1 integrin (Fig. 1) in daughters of irradiated cells was accompanied by increased protein determined by immunoblotting (Fig. 4). In contrast, even though TGF-β treatment decreased E-cadherin and β-catenin protein levels (Fig. 4), localization of E-cadherin and β-catenin immunoreactivity was only affected in double-treated 3D rBM colonies (Fig. 2). Immunostaining can reveal protein access or conformation as well as protein abundance. Preliminary studies suggest that the cell-adhesion proteins of irradiated cells have altered cytoskeletal associations (A.C.E. and M.H.B.-H., unpublished data).
Based on studies in mouse mammary gland, we have proposed that the action of radiation as a carcinogen is augmented by its ability to compromise signaling from the stromal microenvironment (42). A functional test of this concept is provided by our experiments showing that tumorigenesis is increased 4-fold when unirradiated preneoplastic mammary epithelial cells are transplanted to an irradiated mammary stroma (9). One of the most rapid and sensitive responses in the irradiated tissue is the activation of TGF-β (43). TGF-β has a paradoxical effect during carcinogenesis in that it suppresses tumorigenesis but promotes neoplastic progression (44–46). Overexpression of active TGF-β can also induce an epithelial-mesenchymal phenotypic transition during progression in vivo (47). In culture, this phenotype is characterized by loss of E-cadherin, acquisition of mesenchymal cytoskeletal features, and increased cell motility and invasion (48). In our experiments, this effect of TGF-β appears to be augmented by preirradiation of the cells. Similarly, the loss of E-cadherin after very low IR doses may further compromise this essential mediator of cell–cell adhesion in preneoplastic breast cells that already have less E-cadherin (49, 50), and could promote progression.
The loss of cell polarity and multicellular organization exhibited by the progeny of irradiated cells suggest that radiation exposure could promote malignant progression by pathways initially independent of mutational mechanisms. Consistent with this postulate is the observation that colonies from irradiated HMEC contain more cells, indicating that decreased cell–cell communication resulted in loss of contact inhibition and greater proliferation. The events leading to disrupted multicellular organization in the progeny of irradiated HMEC could also contribute to genomic instability. Radiation-induced genomic instability evidenced by increased frequency of mutation and cell death occurs in the progeny of irradiated bone marrow (51, 52) and epithelial cell culture (53). The disruption of cell contacts could permit abnormal cells to persist (54) or dysregulate genome stability functions. Inappropriate mammary expression of an activated metalloprotease in transgenic mice that disrupts cell–ECM interactions and cleaves E-cadherin leads to genomic instability (D. Radisky and M.J.B., unpublished data) and mammary tumors (55, 56).
Here we show that IR can promote phenotypic progression by affecting pathways that inhibit the ability of daughter cells to interact with each other and the microenvironment. Agents designed to protect irradiated tissue from disruption of cell–cell communication (57), or those that can reverse the irradiated phenotype, could provide a means of impeding its downstream carcinogenic potential.
Acknowledgments
We thank Shraddha Ravani for technical assistance and William Chou for preparation of figures. We thank Drs. Joe Gray and Wen-Lin Kuo and the University of California, San Francisco, Cancer Center for CGH analysis and support to C.C.P. This research was supported by the Low Dose Radiation Program, Office of Biological and Environmental Research, Department of Energy (R.L.H.-P. and M.H.B.-H.), National Aeronautics and Space Administration Specialized Center Of Research and Training in Radiation Health (M.H.B.-H. and C.C.P.), Office of Biological and Environmental Research Contract No. DE-AC-03-76SF00098, Department of Energy and Department of Defense/Breast Cancer Research Program Innovator Award (to M.J.B.), and Department of Defense/Breast Cancer Research Program Postdoctoral Training Grant 17-00-0224 (to A.C.E.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: IR, ionizing radiation; TGF-β, transforming growth factor β1; ECM, extracellular matrix; HMEC, human mammary epithelial cells; 3D rBM, 3D reconstituted basement membrane.
References
- 1.Mattsson, A., Ruden, B.-I., Wilking, N. & Rutqvist, L. E. (1993) J. Natl. Cancer Inst. 85, 1679–1685. [DOI] [PubMed] [Google Scholar]
- 2.Mauch, P. (1995) Int. J. Radiat. Oncol. Biol. Phys. 33, 959–960. [DOI] [PubMed] [Google Scholar]
- 3.Davis, F. G., Boice, J. D., Hrubec, Z. & Monson, R. R. (1989) Cancer Res. 49, 6130–6136. [PubMed] [Google Scholar]
- 4.Tokunaga, M., Land, C. E., Yamamoto, T., Asano, M., Tokuoka, S., Ezaki, H. & Nishimori, I. (1987) Radiat. Res. 112, 243–272. [PubMed] [Google Scholar]
- 5.Grosovsky, A. J. (1999) Proc. Natl. Acad. Sci. USA 96, 5346–5347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Trosko, J. E., Chang, C. C. & Madhukar, B. V. (1990) Radiat. Res. 123, 241–251. [PubMed] [Google Scholar]
- 7.Barcellos-Hoff, M. H. (2001) J. Mammary Gland Biol. Neoplasia 6, 213–221. [DOI] [PubMed] [Google Scholar]
- 8.Bissell, M. J. & Radisky, D. (2001) Nat. Rev. Cancer 1, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barcellos-Hoff, M. H. & Ravani, S. A. (2000) Cancer Res. 60, 1254–1260. [PubMed] [Google Scholar]
- 10.Barcellos-Hoff, M. H. (1993) Cancer Res. 53, 3880–3886. [PubMed] [Google Scholar]
- 11.Ehrhart, E. J., Carroll, A., Segarini, P., Tsang, M. L.-S. & Barcellos-Hoff, M. H. (1997) FASEB J. 11, 991–1002. [DOI] [PubMed] [Google Scholar]
- 12.Ewan, K. B., Henshall-Powell, R. L., Ravani, S. A., Pajares, M. J., Arteaga, C., Warters, R., Akhurst, R. J. & Barcellos-Hoff, M. H. (2002) Cancer Res. 62, 5627–5631. [PubMed] [Google Scholar]
- 13.Pierce, G. B., Shikes, R. & Fink, L. M. (1978) Cancer: A Problem of Developmental Biology (Prentice–Hall, Englewood Cliffs, NJ).
- 14.Barcellos-Hoff, M. H., Aggeler, J., Ram, T. G. & Bissell, M. J. (1989) Development (Cambridge, U.K.) 105, 223–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bauer, G. (1996) Histol. Histopathol. 11, 237–255. [PubMed] [Google Scholar]
- 16.Petersen, O. W., Ronnov-Jessen, L., Howlett, A. R. & Bissell, M. J. (1992) Proc. Natl. Acad. Sci. USA 89, 9064–9068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gudjonsson, T., Ronnov-Jessen, L., Billadsen, R., Rank, F., Bissell, M. J. & Petersen, O. W. (2001) J. Cell Sci. 115, 39–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weaver, V. M., Petersen, O. W., Wang, F., Larabell, C. A., Briand, P., Damsky, C. & Bissell, M. J. (1997) J. Cell Biol. 137, 231–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Parvin, B., Yang, Q., Fontenay, G. & Barcellos-Hoff, M. H. (2002) IEEE Comput. 35, 65–71. [Google Scholar]
- 20.Briand, P., Nielsen, K. V., Madsen, M. W. & Petersen, O. W. (1996) Cancer Res. 56, 2039–2044. [PubMed] [Google Scholar]
- 21.Cong, G. & Parvin, B. (2000) in Proceedings of IEEE Conference on Computer Vision and Pattern Recognition, Los Alamitos, CA, Vol. 1, pp. 458–463. [Google Scholar]
- 22.Hodgson, G., Hager, J. H., Volik, S., Hariono, S., Wernick, M., Moore, D., Nowak, N., Albertson, D. G., Pinkel, D., Collins, C., Hanahan, D. & Gray, J. W. (2001) Nat. Genet. 29, 459–464. [DOI] [PubMed] [Google Scholar]
- 23.Weaver, V. M., Lelievre, S., Lakins, J. N., Chrenek, M. A., Jones, J. C., Giancotti, F., Werb, Z. & Bissell, M. J. (2002) Cancer Cell 2, 205–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gumbiner, B. M. (2000) J. Cell Biol. 148, 399–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heimann, R., Lan, F., McBride, R. & Hellman, S. (2000) Cancer Res. 60, 298–304. [PubMed] [Google Scholar]
- 26.Vleminckx, K., Vakaet, L. J., Mareel, M., Fiers, W. & van Roy, F. (1991) Cell 66, 107–119. [DOI] [PubMed] [Google Scholar]
- 27.Yotti, L. P., Trosko, J. E. & Chang, C. C. (1979) Science 206, 1089–1091. [DOI] [PubMed] [Google Scholar]
- 28.Nicolson, G. L., Dulski, K. M. & Trosko, J. E. (1988) Proc. Natl. Acad. Sci. USA 85, 473–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saunders, M. M., Seraj. M. J., Li, Z., SZhou, Z., Winter, C. R., Welch, D. R. & Donahue, H. J. (2001) Cancer Res. 61, 1765–1767. [PubMed] [Google Scholar]
- 30.Jongen, W. M., Fitzgerald, D. J., Asamoto, M., Piccoli, C., Slaga, T. J., Gros, D., Takeichi, M. & Yamasaki, H. (1991) J. Cell Biol. 114, 545–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fujimoto, K., Nagafuchi, A., Tsukita, S., Kuraoka, A., Ohokuma, A. & Shibata, Y. (1997) J. Cell Sci. 110, 311–322. [DOI] [PubMed] [Google Scholar]
- 32.Conacci-Sorrell, M., Zhurinsky, J. & Ben-Ze'ev, A. (2002) J. Clin. Invest. 109, 987–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Luo, J., Lubaroff, D. M. & Hendrix, M. J. (1999) Cancer Res. 59, 3552–3556. [PubMed] [Google Scholar]
- 34.Tlsty, T. D. (2001) Semin. Cancer Biol. 11, 97–104. [DOI] [PubMed] [Google Scholar]
- 35.Rubin, H. (1985) Cancer Res. 45, 2935–2942. [PubMed] [Google Scholar]
- 36.Trosko, J. E. (1998) Environ. Health Perspect. 106, 331–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang, F., Weaver, V. M., Petersen, O. W., Larabell, C. A., Dedhar, S., Briand, P., Lupu, R. & Bissell, M. J. (1998) Proc. Natl. Acad. Sci. USA 95, 14821–14826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang, F., Hansen, R. K., Radisky, D., Yoneda, T., Barcellos-Hoff, M. H., Petersen, O. W., Turley, E. A. & Bissell, M. J. (2002) J. Natl. Cancer Inst. 94, 1494–1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bhatia, R., McGlave, P. B. & Verfaillie, C. M. (1995) J. Clin. Invest. 96, 931–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Akimoto, T., Mitsuhashi, N., Saito, Y., Ebara, T. & Niibe, H. (1998) Int. J. Radiat. Oncol. Biol. Phys. 41, 1171–1176. [DOI] [PubMed] [Google Scholar]
- 41.Meineke, V., Gilbertz, K. P., Schilperoort, K., Cordes, N., Sendler, A., Moede, T. & van Beuningen, D. (2002) Strahlentherapie Onkol. 12, 709–714. [DOI] [PubMed] [Google Scholar]
- 42.Barcellos-Hoff, M. H. (1998) J. Mammary Gland Biol. Neoplasia 3, 165–175. [DOI] [PubMed] [Google Scholar]
- 43.Barcellos-Hoff, M. H., Derynck, R., Tsang, M. L.-S. & Weatherbee, J. A. (1994) J. Clin. Invest. 93, 892–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sieweke, M. H., Thompson, N. L., Sporn, M. B. & Bissell, M. J. (1990) Science 248, 1656–1660. [DOI] [PubMed] [Google Scholar]
- 45.Oft, M., Heider, K.-H. & Beug, H. (1998) Curr. Biol. 8, 1243–1252. [DOI] [PubMed] [Google Scholar]
- 46.Derynck, R., Ackhurst, R. J. & Balmain, A. (2001) Nat. Genet. 29, 117–129. [DOI] [PubMed] [Google Scholar]
- 47.Portella, G., Cumming, S. A., Liddell, J., Cui, W., Ireland, H., Akhurst, R. J. & Balmain, A. (1998) Cell Growth Differ. 9, 393–404. [PubMed] [Google Scholar]
- 48.Janda, E., Lehmann, K., Killisch, I., Jechlinger, M., Herzig, M., Downward, J., Beug, H. & Grunert, S. (2002) J. Cell Biol. 156, 299–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vos, C. B., Cleton-Jansen, A. M., Berx, G., de Leeuw, W. J., ter Haar, N. T., van Roy, F., Cornelisse, C. J., Peterse, J. L. & van de Vijver, M. J. (1997) Br. J. Cancer 76, 1131–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gupta, S. K., Douglas-Jones, A. G., Jasani, B., Morgan, J. M., Pignatelli, M. & Mansel, R. E. (1997) Virchows Arch. 430, 23–28. [DOI] [PubMed] [Google Scholar]
- 51.Kadhim, M. A., Macdonald, D. A., Goodhead, D. T., Lorimore, S. A., Marsden, S. J. & Wright, E. G. (1992) Nature 355, 738–740. [DOI] [PubMed] [Google Scholar]
- 52.Kadhim, M. A., Lorimore, S. A., Townsend, K. M., Goodhead, D. T., Buckle, V. J. & Wright, E. G. (1995) Int. J. Radiat. Biol. 67, 287–293. [DOI] [PubMed] [Google Scholar]
- 53.Mothersill, C., Kadhim, M. A., O'Reilly, S., Papworth, D., Marsden, S. J., Seymour, C. B. & Wright, E. G. (2000) Int. J. Radiat. Biol. 76, 799–806. [DOI] [PubMed] [Google Scholar]
- 54.Barcellos-Hoff, M. H. & Brooks, A. L. (2001) Radiat. Res. 156, 618–627. [DOI] [PubMed] [Google Scholar]
- 55.Lochter, A., Galosy, S., Muschler, J., Freedman, N., Werb, Z. & Bissell, M. J. (1997) J. Cell Biol. 139, 1861–1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sternlicht, M. D., Lochter, A., Sympson, C. J., Huey, B., Rougier, J. P., Gray, J. W., Pinkel, D., Bissell, M. J. & Werb, Z. (1999) Cell 98, 137–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Trosko, J. E. & Ruch, R. (2002) Curr. Drug Targets 3, 465–482. [DOI] [PubMed] [Google Scholar]