Abstract
Terminal deoxynucleotidyl transferase (TdT; EC 2.7.7.31) adds nucleotides to DNA ends generated during V(D)J recombination that are subsequently processed by proteins involved in general double-strand break (DSB) repair pathways. We report an association between TdT and a 55-kDa protein in lymphoid cells. This protein, identified as hPso4, is a homolog of the protein encoded by the PS04/PRP19 gene in Saccharomyces cerevisiae that has pleiotropic functions in DNA recombination and error-prone repair. Purified hPso4 binds double-stranded DNA in a sequence-nonspecific manner but does not bind single-stranded DNA. hPso4 protein is induced 15- to 30-fold in cells by γ radiation and chemical mutagens but not by UV treatment. Loss of hPso4 expression induced by siRNA results in accumulation of DSBs, apoptosis, and decreased cell survival after DNA damage. We conclude that hPso4 plays a major and previously undefined role in mammalian DNA DSB repair.
Keywords: double-strand breaks, end joining
Cells encounter genomic lesions in the form of double-strand breaks (DSBs). In mammalian cells, DSB repair (DSBR) is mediated either by the error-free homologous recombination (HR) or the more predominant error-prone nonhomologous end joining (NHEJ) pathways (1). Agents that induce other types of DNA lesions, such as DNA interstrand crosslinks and adducts that proceed via DSB intermediates, are thought to invoke more than one type of DNA repair, including HR, nucleotide excision repair, and, to a lesser extent, the NHEJ pathway (2). The NHEJ repair proteins Ku70/86, DNA PKcs, and Xrcc4/ligase IV are ubiquitously expressed and function both in generalized DSBR and in the repair of ends generated during V(D)J recombination, a programmed gene rearrangement process that leads to the assembly of Ig and T cell receptors (3). Although these proteins seem sufficient for end joining in vitro, recent studies suggest the requirement of additional unknown factors for end joining in vivo (3, 4). One such recently identified human protein is artemis, mutations in which result in ionizing radiation sensitivity and severe combined immunodeficiency (5). Artemis protein has an inherent 5′ to 3′ single-strand specific exonuclease activity. However, its association with and phosphorylation by DNA PKcs alter its activity to enable endonucleolytic processing of overhangs and cleavage of hairpin substrates such as those generated during V(D)J recombination (6).
Terminal deoxynucleotidyl transferase (TdT; EC 2.7.7.31), a template-independent DNA polymerase, is unique in its expression and function, because it is expressed only in lymphoid cells undergoing V(D)J recombination (3) and has the ability to add nucleotides to the recombining V, (D), and J coding ends, thereby promoting antibody and receptor diversity (7). Our earlier studies revealed the association of TdT with the NHEJ protein Ku via the TdT BRCA1 C terminus (BRCT) region (8). Experiments performed with Ku86-deficient mice and cell lines have indeed demonstrated an absolute requirement of Ku86 for TdT-mediated nucleotide insertions at the V(D)J recombination intermediates (9, 10). The presence of TdT and Ku86 in etoposide-induced nuclear foci (8), the assembly of DNA repair proteins in similar foci after radiation-induced DNA damage (11), and the association of TdT with a large protein complex in thymic nuclear matrix (12) all suggest that TdT may interact with proteins in addition to Ku that are necessary for DSBR.
Our objective was to identify proteins associating with TdT that might participate at DSBs generated during V(D)J recombination and/or DNA repair. This approach has resulted in the identification of hPso4, a homolog of the ScPSO4/PRP19 gene product that functions in DNA repair and recombination (13). Our results suggest that hPso4 has a major previously undefined role in mammalian DNA repair, because its induced deficiency results in apoptosis and decreased clonogenic survival after DNA damage.
Materials and Methods
Cell Lines. All cell lines were obtained from American Type Culture Collection. The origin of the cell lines is as follows: HL60, human leukemia; HeLa, cervical carcinoma; Jurkat, T cell lymphoma; Molt4, preT lymphoma; Nalm6, preB lymphoma; PC12, rat neuronal cells; SV80, human fibroblast; U937, human monocyte; and MRC5, normal human fibroblast. Cells were γ-irradiated by using a Mark I Cs137 or UV-irradiated at a dose of 15 J/m2; drugs used were cisplatin (100 μg/ml), bleomycin (100 μg/ml), mitomycin C (5–50 μg/ml), and etoposide (5–50 μM).
GST Pull-Down. The BRCT domain, N-terminal region of TdT, and TdT were PCR amplified and cloned into pGex-4T-1 (Pharmacia). The expression of the GST-TdT fusion proteins was induced by isopropyl β-D-thiogalactoside. The induced bacterial cell lysate was incubated in buffer containing 150/500 mM NaCl; 50 mM Tris, pH 7.5; 0.3% Triton X-100; 1 mM EDTA; and 1× protease inhibitor (Roche Molecular Biochemicals), with glutathione Sepharose beads for 45 min. The beads were washed (three times), incubated with cell extracts for 2 h, washed again (three times), resuspended in 1.5× loading buffer, electrophoresed on SDS/10% PAGE gels, and stained with Coomassie blue.
Peptide Sequencing. The 55- and 110-kDa polypeptides were excised from Coomassie-stained gels and sequenced by mass spectrometry at the Burroughs Wellcome Facility, Research Triangle Park, NC. The sequence of the 44-kDa TdT was obtained by using the Edman method and starts at STNTGPPKT.
Cloning of Human Pso4. PSO4 cDNA was amplified from Molt-4 polyA+ mRNA with the primers 5′-ATGGGATCCATGTCCCTAATCTGCTCCATCTCT and oligo(dT) (Stratagene). The sequence matched the previously reported sequence for hNMP200 (gi:17391460).
Antibodies. Polyclonal antibodies were raised to the purified GST-Pso4 and affinity purified. Anti-TdT and anti-polyHistidine (His) antibodies were described previously (8). Anti-Cdc5 antibodies were a gift from Peter Groenen (University of Leuven, Leuven, Belgium).
Coimmunoprecipitation Assays. Lysates were processed as described (8). For in vitro binding assays, 1 μg of either purified TdT or hPso4 was mixed in 0.5 ml of binding buffer and processed as described (8).
Purification of Recombinant hPso4. A baculovirus expression construct of human His-Pso4 was generated by using the pFast Bac method (GIBCO-BRL). Recombinant protein was purified from Sf9 cultures and processed per manufacturer's protocol by using Talon affinity beads (Clontech). The fractions were analyzed by SDS/PAGE and Coomassie staining. Fractions containing the purified protein were pooled and dialyzed against 50 mM Tris, pH 7.5/10% glycerol/0.1 mM NaCl/1 mM DTT, and aliquots were stored at –80°C.
Gel Mobility-Shift Assays. Radiolabeled single-stranded (ss)DNA substrate was made by 5′ end labeling with T4 polynucleotide kinase and [γ-32P]ATP. KM1 (5′-GATCTGGCCTGTCTTACACAGTGCTACAGACTGGAACAAAAACCCTGCAG); double-stranded (ds)DNA was made by annealing the labeled KM1 to BO50 (5′-GCAGGGTTTTTGTTCCAGTCTGTAGCACTGTGTAAGACAGGCCAGATCCT). DNA was incubated with varying protein concentrations for 10 min at room temperature in a 10-μl reaction buffer (20 mM Tris, pH 7.5/50 mM NaCl/1 mM DTT/1 mM MgCl2/0.1 mg/ml BSA). Samples were electrophoresed for 60 min at 10 V/cm on a 3.5% polyacrylamide gel in 1× Tris-borate–EDTA buffer.
Small Interfering RNA (siRNA) Transfections. siRNAs were chemically synthesized and ion exchange-HPLC purified to 99.5% purity (Xeragon, Germantown, MD) and are as follows: hPso4 5′-ACCACAGGCUGGCCUCAUUTT-3′, 5′-AAUGAGGCCAGCCUGUGGUTT-3′. Control nonsilencing siRNA was obtained from Xeragon. The annealed ds siRNA was resuspended in buffer (100 mM potassium acetate/30 mM Hepes·KOH/2 mM magnesium acetate, pH 7.4) to obtain a 20 μM solution.
In Situ Labeling of DNA Strand Breaks. siRNA (105) transfected HeLa cells were γ-irradiated (15 Gy) and fixed after 3 h with 1% paraformaldehyde. DNA strand breaks were labeled with Br-dUTP by using TdT, stained with anti-BrdUrd-FITC (BioVision, Mountain View, CA), and detected by flow cytometry (14).
Cell Death ELISA. Assays were set up per manufacturer's instructions with minor modifications (Roche Molecular Biochemicals). Twenty-four hours before transfection, cells were split to 1 × 105 cells per well in a six-well plate and transfected with 2 μg of siRNA by using oligofectamine (Invitrogen). Transfection medium was removed 18–24 h later, and etoposide (5–50 μM) or Miotmycin C (10–50 μg/ml) was added in fresh medium for 1 h. After removal of drug, cells were incubated in fresh medium and harvested after 24 h.
Clonogenic Survival Assays. HeLa cells were transfected with siRNAs as described above. After exposure to DNA-damaging agents, cells were trypsinized, counted, and plated at a density of 200 cells per 100-mm-diameter dish. Twelve to 15 days later, colonies were fixed with 0.2% paraformaldehyde, stained with 0.1% crystal violet, and counted.
Results
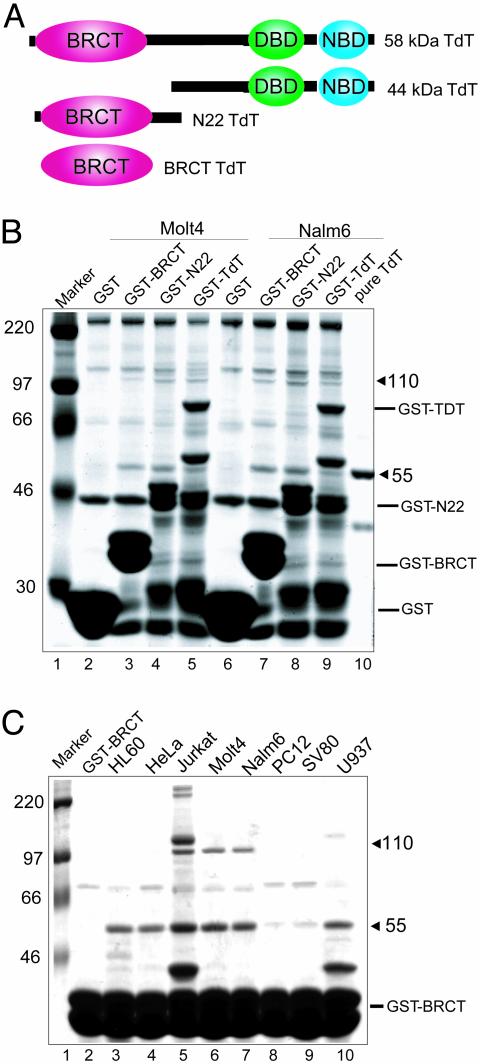
Association Between the BRCT Domain of TdT and a 55-kDa Protein. The BRCT domain found in TdT is a protein–protein interaction motif found in many DNA repair and cell cycle proteins (8). To identify other proteins that might interact with TdT, we generated GST-fusion expression constructs encoding the BRCT domain of TdT alone, the N-terminal fragment of TdT containing the BRCT domain but lacking the catalytic domain, or full-length TdT (Fig. 1A). Whole cell extracts prepared in buffers containing 500 mM NaCl from Molt-4 and Nalm-6 lymphoid cells were incubated with the GST-fusion proteins prebound to glutathione Sepharose beads. Two proteins of 55 and 110 kDa were specifically isolated with all three fusion proteins but not with GST alone (Fig. 1B, compare lanes 2 and 6 to lanes 3–5 and 7–9). The interaction of TdT with the two proteins was also observed at physiological salt concentration (150 mM) and occurred in all cell lines examined, whereas the interaction with the 110-kDa protein was restricted to lymphoid cell lines (Fig. 1C, lanes 5–7). Because these interactions are present at high salt concentration (500 mM NaCl), they represent the specific interaction of each protein with the BRCT domain of TdT.
Fig. 1.
Association of TdT with 55- and 110-kDa proteins. (A) Schematic representation of TdT constructs. Full-length TdT (58 kDa) containing an N-terminal BRCT domain, a DNA-binding domain (DBD), and a nucleotide-binding domain (NBD); 44-kDa protein lacking the BRCT domain; 22-kDa protein lacking the DBD and NBD domains; BRCT domain only. (B and C) Proteins were resolved on SDS/10% PAGE gels and visualized by Coomassie staining. (B) GST pull-down of Molt-4 and Nalm-6 proteins binding to TdT constructs. Lane 1, molecular weight markers; lanes 2–5, GST or GST-fusion proteins incubated with 500 μg of Molt-4 cell extract; lanes 6–9, GST or GST-fusion proteins incubated with 500 μg of Nalm-6 cell extract; lane 10, purified TdT. (C) Binding of proteins from indicated tumor cell lines to the GST-BRCT. Five hundred micrograms of extracts were incubated with GST-BRCT protein in buffers containing 500 mM NaCl and 0.3% Triton X-100. Lane 1, molecular weight markers; lane 2, GST-BRCT alone; lanes 3–10, whole cell extracts incubated with fusion protein.
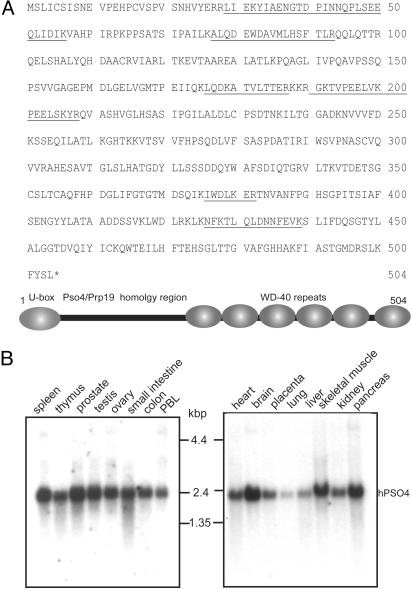
Microsequencing and Characterization of the 55-kDa Protein. To further characterize the TdT-interacting proteins, gel slices containing the 55- and 110-kDa proteins were microsequenced. Mass spectrometric analysis revealed the sequences of six polypeptides from the 55-kDa protein (Fig. 2A). blast search of the protein database demonstrated that one peptide fragment was identical to a peptide encoded by a 200-bp human-expressed sequence tag. RT-PCR from the Molt-4 cell line amplified a 1.6-kb DNA fragment that encoded a protein that contained all of the six polypeptide fragments (Fig. 2A). Northern analysis showed ubiquitous expression of the corresponding 2.4-kb mRNA in human multiple tissue blots (Clontech) (Fig. 2B). A protein blast search revealed 38% sequence identity and 56% similarity with the ScPSO4/PRP19 gene product, the highest degree of homology observed for the 55-kDa protein in yeast, and >80% homology with an uncharacterized ≈55-kDa protein from rodents and Drosophila (Fig. 8, which is published as supporting information on the PNAS web site, www.pnas.org). We therefore refer to this protein as hPso4. Domain analysis predicts similar protein architecture among plant, yeast, rodent, and human proteins. The U box at the N terminus is a modified RING finger domain present in a subset of ubiquitin ligases such as RAD5 that has been hypothesized to have a role in E2-binding and ubiquitination of the target protein in response to DNA damage (15). The C terminus of hPso4 contains six successive WD-40 or β transducin-like repeats (Fig. 2A), which are known to form a structural interface for the assembly of multiprotein complexes. hPso4 has also been identified as a component of the nuclear matrix (NMP200) (16).
Fig. 2.
Sequence and schematic representation of the 55-kDa protein. (AUpper) Amino acid sequences of the hPso4 peptide fragments identified by mass spectrometry are underlined. (Lower) Schematic representation of the hPso4 protein showing the U-box location, Prp19 homology region, and WD-40 repeats. (B) Northern analysis of hPSO4 mRNA in human multiple tissue blots (Clontech).
Peptide analysis of the 110-kDa protein revealed it to be a previously identified cell division cycle protein, hCdc5 (17). Western analysis showed ubiquitous expression of hCdc5 in most cell lines (data not shown) in agreement with the hCdc5 mRNA expression data from several adult tissues (17). However, hCdc5 was pulled down with TdT fusion proteins only in extracts from cell lines expressing endogenous TdT and hPso4, whereas hPso4 associated with TdT in all cell lysates tested. This result suggests that the interaction between hCdc5 and TdT is indirect, possibly requiring additional protein(s) present only in lymphoid cells.
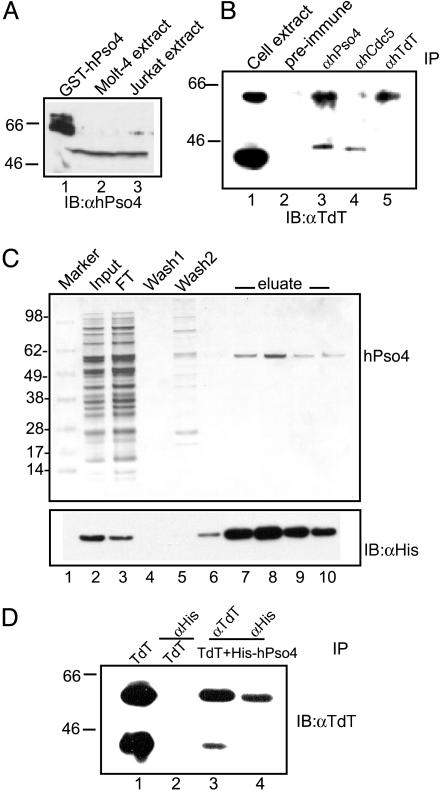
Interaction of hPso4 with TdT in Vivo and in Vitro. To determine whether the interaction between hPso4 and TdT is physiologically relevant, we performed coimmunoprecipitation experiments in lymphoid cell extracts. Whole cell extracts were prepared from Molt-4 cells that contain endogenous TdT and hPso4. Lysates were incubated with either rabbit preimmune serum, anti-hPso4, anti-hCdc5, or anti-TdT antibodies. The immunoprecipitates were tested for the presence of TdT. Anti-TdT and anti-hPso4 antibodies immunoprecipitated the 58-kDa TdT, whereas the preimmune serum and anti-hCdc5 did not (Fig. 3B). Anti-hPso4 and anti-hCdc5 antibodies also immunoprecipitated a small amount of the 44-kDa TdT present in the cell lysate (Fig. 3B, lanes 1, 3, and 4). Coimmunoprecipitations were performed at physiological salt concentration (150 mM NaCl), a condition permissive for DNA–protein interactions. Because the 44-kDa TdT, 58-kDa TdT, and hCdc5 bind DNA, their presence in hPso4 and hCdc5 antibody pull-downs could be due to indirect interactions mediated by DNA.
Fig. 3.
Coimmunoprecipitation of TdT and hPso4. (A) Western blots of 50 μg of Molt-4 and Jurkat cell extracts by using affinity-purified rabbit polyclonal antibodies raised to the GST-Pso4 fusion protein. (B) Immunoprecipitation of TdT from 200μg of Molt-4 extract by using preimmune serum, anti-hPso4, anti-hCdc5, and anti-TdT antibodies. Immune complexes were probed by using anti-TdT antibody. Lane 1 contains 60 μg of Molt4 lysate. (C) Purification of recombinant hPso4 from Sf9 cells. (Upper) Coomassie stain. Lane 1, molecular weight markers; lane 2, input; lane 3, FT flow-through; lanes 4 and 5, wash; lanes 6–10, 100 mM imidazole eluates. (Lower) Western blot by using anti-His antibodies. (D) Interaction of TdT and hPso4. Lane 1, purified TdT; lane 2, purified TdT with anti-His antibody; 58-kDa TdT immunoprecipitated from a preformed complex of 44-kDa TdT, 58-kDa TdT, and His-hPso4 with anti-TdT (lane 3) or anti-His antibody (lane 4).
To eliminate such potential indirect associations, coimmunoprecipitations were performed with purified proteins. To obtain purified hPso4, a baculovirus construct was generated with a six-His tag at the N terminus of hPso4. The protein was purified to >99% homogeneity from Sf9 insect cells on a Talon affinity column (Fig. 3C Upper, lanes 7–10). Anti-His antibodies reconfirmed the purification of hPso4 (Fig. 3C Lower, lanes 7–10). Purification of TdT has been reported (8). Coimmunoprecipitation assays were performed with a mixture of purified proteins containing the 58-kDa TdT, 44-kDa TdT, and His-hPso4 (Fig. 3D). Anti-His antibodies coimmunoprecipitated only the 58-kDa TdT and not the 44-kDa TdT from the mixture (Fig. 3D; compare lanes 2, 3, and 4), indicating that only the 58-kDa TdT physically associates with hPso4. Because the 44-kDa TdT lacks the BRCT domain of TdT (Fig. 1 A), it appears that the BRCT domain is essential for the interaction between TdT and hPso4.
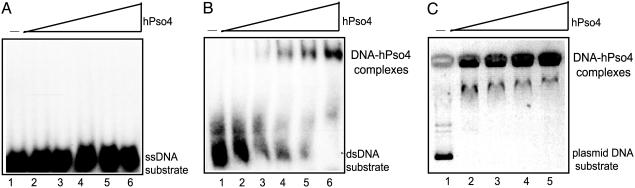
DNA-Binding Properties of hPso4. TdT catalyzes nucleotide polymerization at DNA ends. We therefore tested whether purified hPso4 binds directly to DNA by using gel mobility-shift assays and radiolabeled ss- and dsDNA substrates. hPso4 did not bind 32P-labeled 50-mer ss oligo (5 nM) over a range of protein concentrations (90–540 nM) (Fig. 4A, lanes 2–6). In contrast, a single high-mobility band was observed on incubation of increasing concentrations (90–540 nM) of hPso4 with 32P-labeled 50-bp dsDNA (5 nM) (Fig. 4B, lanes2–6). hPso4 also readily bound plasmid DNA substrates, suggesting a lack of sequence specificity. DNA–protein complexes were also observed with closed circular plasmid DNA, indicating that hPso4 does not require free ends to bind DNA (Fig. 4C, lanes 2–5).
Fig. 4.
Binding of hPso4 to DNA radiolabeled DNA was incubated with purified hPso4 protein. The reaction products were analyzed on native polyacrylamide gels followed by autoradiography (A and B) or 0.7% agarose gel stained with ethidium bromide (C). (A) hPso4 binding to ssDNA. Lane 1, 32P-labeled 50-mer ssDNA (5 nM) and increasing amounts of hPso4 (lanes 2–6, 90–540 nM). (B) hPso4 binding to dsDNA; 5 nM 32P-labeled 50 bp of duplex DNA (lane 1) and increasing amounts of hPso4 (lanes 2–6, 90–540 nM). (C) hPso4 binding to circular plasmid DNA; 12 nM pBKS dsDNA (lane 1) and increasing amounts of hPso4 (lanes 2–5, 0.4–0.7 μm).
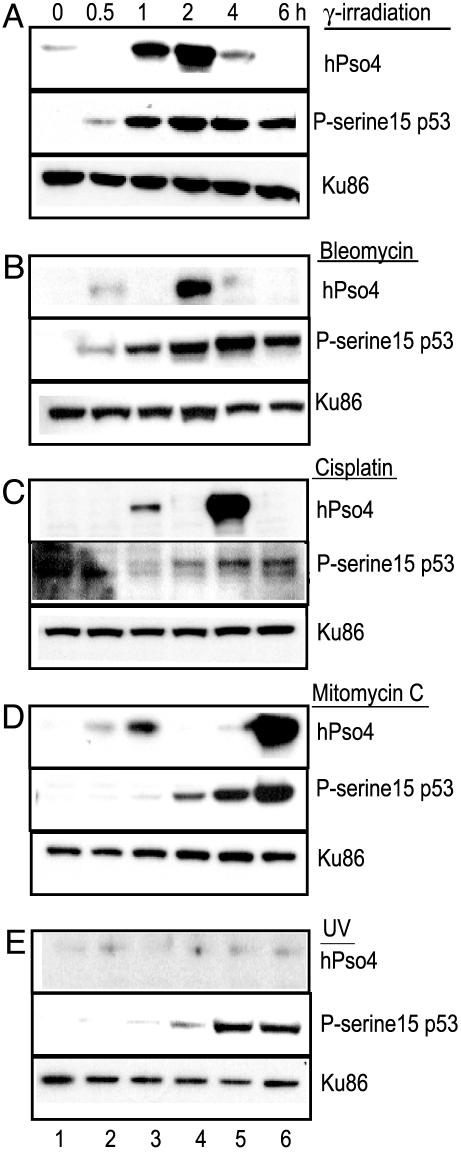
Up-Regulation of hPso4 After DNA Damage. In view of the ability of hPso4 to bind dsDNA in a sequence-nonspecific manner and the role of ScPSO4 gene product in DNA end joining (18), we examined the role of hPso4 in mammalian DNA repair by exposing cells to genotoxic agents that create lesions in the DNA. The normal human fibroblast cell line, MRC5, was γ-irradiated, and whole cell extracts were prepared at different times postirradiation. Western analysis demonstrated a 17- to 30-fold increase in hPso4 protein levels at 1–2 h, followed by a rapid decrease by 4 h (Fig. 5A, lanes 3–6). The increase in hPso4 expression was concurrent with the phosphorylation of p53 at Ser-15, an event that signals DNA damage (Fig. 5A Middle). Levels of the NHEJ protein Ku86 were not altered after γ irradiation (Fig. 5A Bottom). A similar induction of hPso4 levels after irradiation exposure was also observed in two unrelated fibroblast and lymphoid cell lines (data not shown).
Fig. 5.
Time course of hPso4 induction in response to DNA damage MRC5 cells treated with 12-Gy γ irradiation (A), 100 μg/ml bleomycin (B), 100 μg/ml cisplatin (C), 50 μg/ml mitomycin C (D), and 15 J/m2 uv (E). Western blots containing 30 μg of cell extracts prepared from untreated or treated (0.5–6 h) cells were probed with anti-hPso4, antiphosphoserine15-p53 (P-serine 15), or anti-Ku86 antibodies, as indicated.
HPso4 was also up-regulated in response to the antitumor DSB-inducing agent bleomycin (Fig. 5B), whereas the DNA crosslinking agents cisplatin (Fig. 5C) and mitomycin C (Fig. 5D) caused biphasic induction in hPso4 levels. The initial 2.9- to 3.5-fold induction at 1 h could be duetothe crosslinks that are first formed in the DNA, whereas the 15-fold increases at 4 and 6 h, respectively, coincide with the maximal intensity of p53 Ser-15 phosphorylation, suggesting the appearance of DSBs at these time points. In contrast, cells treated with UV radiation (15 J/m2) did not demonstrate induction (Fig. 5E).
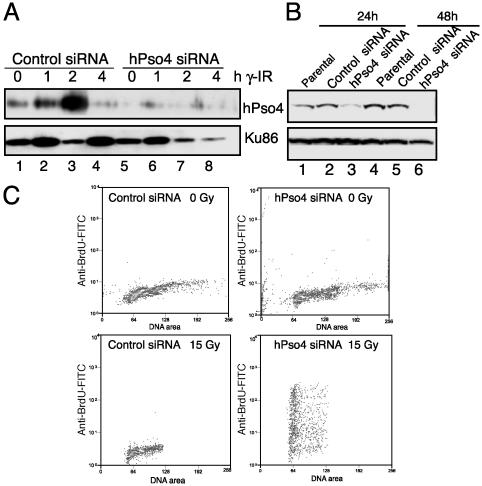
Effects of Down-Regulation of hPso4 by siRNA on DSBs, Apoptosis, and Cell Survival After DNA Damage. To determine whether the induction of hPso4 is essential for DNA repair in vivo, we analyzed the response of hPso4-deficient cells to DNA damage. To this end, we synthesized siRNAs directed to an internal region of hPSO4 cDNA. MRC5 cell lines transfected with hPso4 siRNA but not the control siRNA specifically down-regulate the levels of hPso4 protein and prevent further induction after 12 Gy of γ irradiation (Fig. 6A). Although the overall protein loading is not uniform, as judged by the amount seen on Ku Western blots, the protein levels at 0, 1, and 2 h are internally comparable between the control and hPso4 siRNA transfected cells. Similarly, HeLa cells transfected with hPso4 siRNA demonstrate significant reduction in hPso4 protein levels 24–48 h posttransfection (Fig. 6B).
Fig. 6.
Effect of siRNA transfection on hPso4 expression. (A) MRC5 cells were transfected with control or hPso4 siRNA. Twenty-four hours posttransfection, cells were γ-irradiated at 12 Gy. Western blots containing 75 μg of cell extracts prepared from nonirradiated or irradiated (1–4 h) cells were probed with anti-hPso4 or anti-Ku86 antibodies, as indicated. (B) Western analysis of whole cell extracts prepared from HeLa cells (1.5 × 106) transfected with either the control siRNA (lanes 2 and 5) or the hPso4-siRNA (lanes 3 and 6) and probed with anti-hPso4 or anti-Ku 86 antibodies. (C) Flow cytometry of control and hPso4 siRNA transfected cells unirradiated (Upper) or γ-irradiated at 15 Gy (Lower), incubated in fresh growth medium to allow repair, and fixed after 3 h. Cells were stained with anti-BrdUrd-FITC to detect DSBs at which Br-dUTP had been incorporated by using TdT (14, 19). Similar results were obtained in a second experiment.
To test whether hPso4 is necessary for the repair of DSBs, cells transfected with control or hPso4 siRNA for 24 h were then exposed to 15 Gy γ irradiation followed by repair incubation for 3 h and fixed (19). DSBs were labeled in situ with Br-dUTP by using TdT, stained with anti-BrdUrd-FITC, and detected by flow cytometry (14). Cells transfected with hPso4 siRNA demonstrated ≈100-fold increase in Br-dUTP incorporation after DNA damage (Fig. 6C) in two independent transfections, indicative of the presence of unrepaired strand breaks, whereas the anti-BrdUrd-FITC staining of irradiation-exposed control siRNA cells is near the levels seen in unirradiated cells, suggesting the repair of genotoxic lesions.
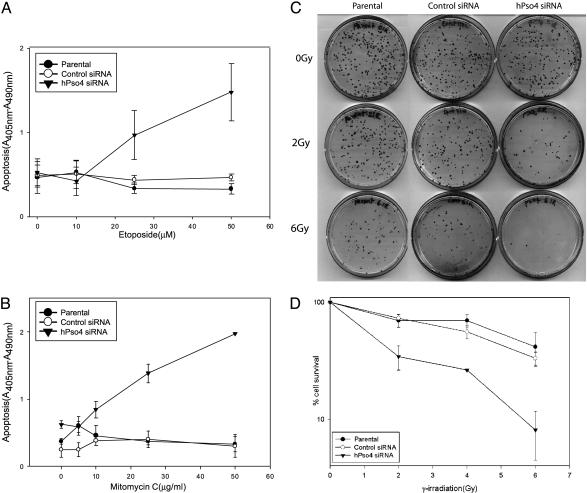
To determine whether the increase in DSBs in hPso4 siRNA transfected cells correlated with a decrease in cell survival, cells transfected with the siRNAs were treated with etoposide (10–50 μM) or mitomycin C (5–50 μg/ml) and assayed for apoptosis after 24 h. hPso4-deficient cells underwent rapid dose-dependent apoptotic cell death with both DNA-damaging agents, whereas the parental and the control siRNA transfected cells did not (Fig. 7 A and B). Clonogenic survival assays also showed a significant decrease (P < 0.05) in cell survival of the hPso4-deficient cells, with increasing doses of both γ irradiation (Fig. 7 C and D) and etoposide (data not shown) in comparison to the parental cells or the control siRNA transfected cells, strongly suggesting defective DNA repair in the hPso4-deficient cells.
Fig. 7.
Effect of hPso4 on cell survival after DNA damage. Cell death ELISAs were performed on 105 HeLa cells or cells transfected with the control or hPso4 siRNAs and treated with etoposide (A) or mitomycin C (B) for 1 h. Each graph is the result of three independent transfections assayed in duplicate and represents the mean ± SEM. Clonogenic survival assays were performed on 105 HeLa parental cells or cells transfected with the control or hPso4 siRNAs and exposed to indicated doses of γ irradiation. Cells were trypsinized and then plated at a density of 200 cells per 100-mm-diameter dish. Surviving colonies were counted after 12 days.(C) A representative clonogenic survival assay. (D) Survival data from three independent transfections assayed in triplicate. Data are shown as percent of control and represent the mean ± SEM.
Discussion
We have identified a human homolog of the yeast Pso4 protein through its association with the lymphoid-specific polymerase, TdT. The hPso4 protein is ubiquitously expressed, and its expression is increased in response to γ irradiation, radiomimetic drugs, and DNA crosslinking agents. Furthermore, decreased expression of the protein induced by siRNA before and after DNA damage leads to accumulation of DSBs, marked enhancement of apoptosis, and decreased clonogenic survival of HeLa cells expressing endogenous hPso4 protein. These observations strongly support the hypothesis that hPso4 plays an important role in DNA repair processes that result from the induction of DSBs.
Studies in yeast have led to the identification of nine distinct structurally unrelated genes important for resistance to the lethal effects of psoralen-UVA induced DNA crosslinks. Of these “PSO” genes, seven have been demonstrated to have a role in DNA repair. Notably, in pso2-1/snm1 and pso4-1 mutants, accumulation of DSBs was found by sucrose gradient analysis after interstrand crosslink-induced DNA damage, implicating their gene products in the processing and/or rejoining of DNA ends (18, 20, 21). Genetic interactions have also been observed among yeast pso mutants, suggesting that their gene products may functionally interact to repair crosslink-induced DNA damage (18, 20).
Members of the mammalian Pso2/Snm1 family have also been documented to play important roles in DNA repair. Mouse snm1(–/–) embryonic stem cell lines and snm1 knockout mice are sensitive to mitomycin C (22). hSnm1 is a human ScPSO2/SNM1 homolog (22, 23) that shows a 4- to-5-fold increase in nuclear foci after exposure to irradiation or DNA crosslinking agents and colocalizes with the p53-binding protein 53BP1 before and after irradiation and with hMre11 after irradiation, suggesting it is recruited to repair foci (23). Patients with mutations in the Snm1 homology domain of Artemis (SNM1C) show hypersensitivity to ionizing radiation and V(D)J recombination defects (5, 24). Our finding that hPso4 is essential for the optimal repair of DSBs further underscores the importance of this group of proteins in mammalian cells and will lead to further studies to define the molecular mechanisms by which they act at DNA ends.
Under stringent conditions, TdT stably associates with hPso4 through its BRCT domain in several cell lines. The BRCT domain is also present in the NHEJ proteins pol μ and Xrcc4 that assemble as multiprotein complexes at DNA ends to assist in end joining (25) and in the DSBR protein Nbs1 that binds γ-H2AX through its fork-head associated/BRCT domain. This domain is also necessary for the formation of hMRE11/RAD50/Nbs1 foci and cell survival after irradiation (26). The interaction of hPso4 with TdT through the BRCT domain suggests that hPso4 may be recruited to repair sites through protein–protein interactions and perhaps in turn stabilizes the repair complexes assembled at these sites by way of its WD-40 repeats, which are known to have a scaffolding role. Future studies will determine whether these repeats are important for the function of hPso4 in DSBR.
Supplementary Material
Acknowledgments
We thank Dr. Will Burkhart and Kevin Blackburn at Burroughs Wellcome for protein sequencing, Nancy Martin for FACS analysis, and Dr. Peter Groenen for hCdc5 antibodies. We also thank Drs. Nupam Mahajan, Josef Spychala, and Dale Ramsden for critical reading of the manuscript and Dr. David Sorscher for suggestions. K.N.M is a recipient of the Ruth Kirschtein Post Doctoral award. This work was supported by Public Health Service Grant RO1 CA34085 (to B.S.M.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: DSB, double-strand break; TdT, terminal deoxynucleotidyl transferase; NHEJ, nonhomologous end joining; DSBR, DSB repair; BRCT, BRCA1 C terminus; His, histidine; siRNA, small interfering RNA; ds, double-stranded; ss, single-stranded.
References
- 1.Jackson, S. P. (2002) Carcinogenesis 23, 687–696. [DOI] [PubMed] [Google Scholar]
- 2.D'Andrea, A. D. & Grompe, M. (2003) Nat. Rev. Cancer 1, 23–34. [DOI] [PubMed] [Google Scholar]
- 3.Gellert, M. (2002) Annu. Rev. Biochem. 71, 101–132. [DOI] [PubMed] [Google Scholar]
- 4.Dai, Y., Kysela, B., Hanakahi, L. A., Manolis, K., Riballo, E., Stumm, M., Harville, T. O., West, S. C., Oettinger, M. A. & Jeggo, P. (2003) Proc. Natl. Acad. Sci. USA 5, 2462–2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li, L., Moshous, D., Zhou, Y., Wang, J., Xie, G., Salido, E., Hu, D., de Villartay, J. P. & Cowan, M. J. (2002) J. Immunol. 168, 6323–6329. [DOI] [PubMed] [Google Scholar]
- 6.Ma, Y., Pannicke, U., Schwarz, K. & Lieber, M. R. (2002) Cell 108, 781–794. [DOI] [PubMed] [Google Scholar]
- 7.Benedict, C. L., Gilfillan, S., Thai, T. H. & Kearney, J. F. (2000) Immunol. Rev. 175, 150–157. [PubMed] [Google Scholar]
- 8.Mahajan, K. N., Gangi-Peterson, L., Sorscher, D. H., Wang, J., Gathy, K. N., Mahajan, N. P., Reeves, W. H. & Mitchell, B. S. (1999) Proc. Natl. Acad. Sci. USA 96, 13926–13931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bogue, M. A., Wang, C., Zhu, C. & Roth, D. B. (1997) Immunity 7, 37–47. [DOI] [PubMed] [Google Scholar]
- 10.Purugganan, M. M., Shah, S., Kearney, J. F. & Roth, D. B. (2001) Nucleic Acids Res. 29, 1638–1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bradbury, J. M. & Jackson, S. P. (2003) Biochem. Soc. Trans. 31, 40–44. [DOI] [PubMed] [Google Scholar]
- 12.Pandey, V. N., Dave, V. P., Patil, M. S., Pradhan, D. S., Amrute, S. B. & Modak, M. J. (1990) Biochemistry 29, 4037–4041. [DOI] [PubMed] [Google Scholar]
- 13.Grey, M., Dusterhoft, A., Henriques, J. A. & Brendel, M. (1996) Nucleic Acids Res. 24, 4009–4014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li, X. & Darzynkiewicz, Z. (1995) Cell Prolif. 28, 571–579. [DOI] [PubMed] [Google Scholar]
- 15.Ulrich, H. D. (2003) J. Biol. Chem. 278, 7051–7058. [DOI] [PubMed] [Google Scholar]
- 16.Gerner, C., Holzmann, K., Meissner, M., Gotzmann, J., Grimm, R. & Sauermann, G. (1999) J. Cell Biochem. 74, 145–151. [PubMed] [Google Scholar]
- 17.Bernstein, H. S. & Coughlin, S. R. (1997) J. Biol. Chem. 272, 5833–5837. [DOI] [PubMed] [Google Scholar]
- 18.de Morais, M. A., Jr., Vicente, E. J., Brozmanova, J., Schenberg, A. C. & Henriques, J. A. (1996) Curr. Genet. 29, 211–218. [DOI] [PubMed] [Google Scholar]
- 19.Kodym, R. & Horth, E. (1995) Int. J. Radiat. Biol. 68, 133–139. [DOI] [PubMed] [Google Scholar]
- 20.Bredel, M. & Henriques, J. A. (2001) Mutat. Res. 489, 79–96. [DOI] [PubMed] [Google Scholar]
- 21.Magana-Schwencke, N., Henriques, J. A., Chanet, R. & Moustacchi, E. (1982) Proc. Natl. Acad. Sci. USA 79, 1722–1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dronkert, M. L., de Wit, J., Boeve, M., Vasconcelos, M. L., van Steeg, H., Tan, T. L., Hoeijmakersm, J. H. & Kanaar, R. (2000) Mol. Cell. Biol. 20, 4553–4561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Richie, C. T., Peterson, C., Lu, T., Hittelman, W. N., Carpenter, P. B. & Legerski, R. J. (2002) Mol. Cell. Biol. 20, 8635–8647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Noordzij, J. G., Verkaik, N. S., van der Burg, M., van Veelen, L. R., de Bruin-Versteeg, S., Wiegant, W., Vossen, J. M., Weemaes, C. M., de Groot, R., Zdzienicka, M. Z., et al. (2003) Blood 101, 1446–1452. [DOI] [PubMed] [Google Scholar]
- 25.Mahajan, K. N., Nick McElhinny, S. A., Mitchell, B. S. & Ramsden, D. A. (2002) Mol. Cell. Biol. 14, 5194–5202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhao, S., Renthal, W. & Lee, E. Y. (2002) Nucleic Acids Res. 30, 4815–4822. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.