Abstract
Structural protein 4.1, which has crucial interactions within the spectrin–actin lattice of the human red cell membrane skeleton, also is widely distributed at diverse intracellular sites in nucleated cells. We previously showed that 4.1 is essential for assembly of functional nuclei in vitro and that the capacity of 4.1 to bind actin is required. Here we report that 4.1 and actin colocalize in mammalian cell nuclei using fluorescence microscopy and, by higher-resolution detergent-extracted cell whole-mount electron microscopy, are associated on nuclear filaments. We also devised a cell-free assay using Xenopus egg extract containing fluorescent actin to follow actin during nuclear assembly. By directly imaging actin under nonperturbing conditions, the total nuclear actin population is retained and visualized in situ relative to intact chromatin. We detected actin initially when chromatin and nuclear pores began assembling. As nuclear lamina assembled, but preceding DNA synthesis, actin distributed in a reticulated pattern throughout the nucleus. Protein 4.1 epitopes also were detected when actin began to accumulate in nuclei, producing a diffuse coincident pattern. As nuclei matured, actin was detected both coincident with and also independent of 4.1 epitopes. To test whether acquisition of nuclear actin is required for nuclear assembly, the actin inhibitor latrunculin A was added to Xenopus egg extracts during nuclear assembly. Latrunculin A strongly perturbed nuclear assembly and produced distorted nuclear structures containing neither actin nor protein 4.1. Our results suggest that actin as well as 4.1 is necessary for nuclear assembly and that 4.1–actin interactions may be critical.
We are investigating functions of the protein 4.1 family in nuclei, centrosomes, and mitotic spindles in eukaryotic cells in relation to cell division (1–3). Protein 4.1 is a superfamily of multifunctional structural proteins widely expressed in nucleated cells (4), the prototypical member of which has well defined interactions with spectrin, actin, and integral membrane proteins in the human red cell membrane skeleton (reviewed by refs. 5 and 6). We recently showed that protein 4.1 is essential for proper assembly of functional nuclei in vitro in Xenopus egg extracts and identified the 4.1 spectrin–actin binding domain (SABD) as one of the 4.1 domains critical for this process. Furthermore, using a mutant 4.1 SABD unable to bind actin, we demonstrated that 4.1–actin binding capacity is necessary for nuclear reconstitution (3). These observations prompted us to investigate further the roles of actin in nuclear assembly.
Actin is widely recognized as a major cytoskeletal component involved in dynamic processes such as cell motility and shape changes, cytoplasmic vesicle transport, and muscle contraction. Actin also was reported for several decades to be intranuclear, in particular to be associated with nuclear matrix (7–9), although many of these reports initially met with skepticism. Objections included possible cytoplasmic contamination, extraction and fixation artifacts, and the inability to detect nuclear actin by using fluorescently labeled phalloidin. However, recent experimental data confirm and extend the earlier reports to convincingly provide evidence documenting actin in nuclei. These newer observations include characterization of actin and actin-related proteins in chromatin remodeling and histone acetyl transferase complexes (reviewed by ref. 10) and identification of two functional leucine-rich nuclear export signals in actin (11). In addition to protein 4.1, other actin-binding proteins in nuclei have been identified (reviewed by ref. 12) including a nuclear-specific myosin (13), a nuclear-specific spectrin (14), and cofilin (15).
We report here that 4.1 and actin colocalize in nuclei of cultured mammalian cells by fluorescence microscopy of fixed and live cells. Higher-resolution electron microscopy (EM) of detergent-extracted cell whole mounts showed actin and 4.1 epitopes closely associated on nuclear filaments. However, we also imaged nuclear actin directly during nuclear assembly in vitro by using Xenopus egg extracts and fluorescently labeled actin. Under nonperturbing conditions, we initially detected actin during nuclear reconstitution when chromatin and nuclear pores began to assemble. A discrete nuclear actin network-like pattern formed as nuclear lamina assembled but before DNA synthesis. We also report that actin acquisition is necessary for proper nuclear assembly, and inhibition of either actin or 4.1 excludes both actin and protein 4.1 from being incorporated into nuclei, blocking nuclear assembly.
Methods
Materials. WI38 cells (CCL 75) were obtained from the American Type Culture Collection. BrdUrd was from Sigma, enhanced yellow fluorescent protein (EYFP) and enhanced cyan fluorescent protein (ECFP) vectors were from CLONTECH, and lipofectamine was from Invitrogen. Rhodamine-labeled actin was the kind gift of M. Welch (University of California, Berkeley). Latrunculin A was generously provided by D. Drubin (University of California, Berkeley), and anti-lamin was from A. Merdes (University of Edinburgh, Edinburgh). IgGs against 4.1R SABD and 4.1R C-terminal domain have been described (1) and were kindly provided by J. Chasis (Lawrence Berkeley National Laboratory, Berkeley). Antibodies against BrdUrd were from BD Biosciences, mAb 414 against nuclear pore complex proteins was from Babco (Richmond, CA), mAb C4 against actin was from ICN, secondary fluorescent antibodies were from Molecular Probes, and gold-bead-conjugated secondary antibodies were from Amersham Pharmacia.
Xenopus Extracts and Nuclear Assembly Reactions. Cytoplasmic Xenopus egg extracts (10,000 × g) were prepared as described (16) and made interphasic with 400 mM Ca2+. For nuclear assembly, demembranated Xenopus sperm were added to 20 μl of egg extract on ice with rhodamine-labeled actin and incubated at 20°C. Assembly reactions were imaged directly in squashes of reaction aliquots and Fix (16) for convenience, although nuclear actin was also visible in unfixed samples. For immunostaining, reactions diluted with BRB80 (80 mM Pipes/2 mM MgCl2/1 mM EGTA, pH 6.8)/15% glycerol/1% Triton X-100 were spun through 25% glycerol-BRB80 cushions onto coverslips, fixed in –20°C methanol, and probed with antibodies as described (3). Images were captured by using a Nikon Eclipse 2000 microscope equipped with a CCD camera and processed by using photoshop (Adobe Systems, Mountain View, CA). Western blotting of nuclei and BrdUrd labeling were done as described (3).
Immunofluorescent Microscopy of Human Diploid Fibroblasts. Growth of WI38 cells, nuclear matrix preparation, immunofluorescent staining, and transient transfections were performed as described (1). Controls with equal amounts of nonimmune IgG, no primary antibody, or vector alone were included in each experiment as appropriate.
Whole-Mount Embedment-Free Immuno-EM. High-resolution whole-mount EM of detergent-extracted, immunostained WI38 diploid human fibroblasts was performed as described (17). Briefly, cells grown on parlodion-carbon-coated sterile nickel grids were extracted, fixed with paraformaldehyde/glutaraldehyde, blocked with goat serum, incubated with primary antibody, and immunostained with gold-bead-conjugated secondary antibodies. Samples were fixed again with glutaraldehyde, washed in 0.1 M cacodylate (pH 7.4), dehydrated in ethanol, and critical point-dried. Sample grids thinly coated with carbon were viewed by using a JEOL 1200EX electron microscope with a total tilt angle of 10° for stereo.
Results
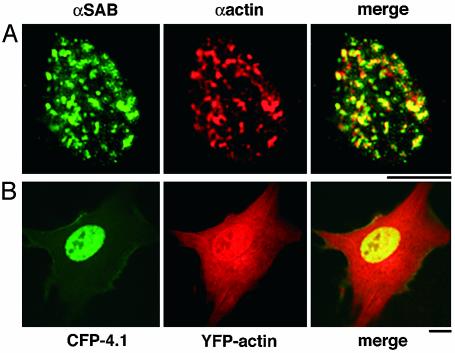
Localization of Protein 4.1 and Actin in Nuclei. Actin can be detected in nuclei of mammalian cells by immunofluorescence microscopy after cells are first extracted with detergent and high salt and then DNA is removed by enzymatic digestion to reveal the nuclear matrix or scaffold. Previously, actin and protein 4.1 isoforms were detected independently in nuclear matrix by immunofluorescence microscopy and Western blot analysis in a variety of cell types (1, 8, 9, 18–20). Therefore, to localize 4.1 relative to nuclear actin, we extracted cultured mammalian cells and probed nuclear matrix by double-label immunofluorescence (Fig. 1A). Anti-actin produced a punctate pattern along with a more diffuse staining in nuclei. The punctate immunofluorescent labeling by protein 4.1 antibody was largely coincident with focal anti-actin immunofluorescent signals at this level of resolution.
Fig. 1.
Colocalization of nuclear actin and 4.1 in human diploid fibroblasts. (A) Double-label immunofluorescence of 4.1 SABD epitopes (green) and actin (red) in WI38 cells extracted in situ to prepare nuclear matrix. The merged image shows a high degree of coincidence (yellow). (B) Transient cotransfection of WI38 cells with YFP-actin (red) and CFP-80kD4.1R (green). Cells were imaged 4–72 h posttransfection. The 24-h image presented shows expression of YFP-actin in both cytoplasm and nucleus, whereas CFP-4.1 is exclusively nuclear, generating coincident nuclear signals (yellow). (Bar, 10 μm.)
In a second independent approach, actin and 4.1 proteins with fluorescent tags were expressed in live cells. Vectors with CFP fused to actin or YFP fused to 80-kDa protein 4.1R containing an SABD were transfected into WI38 diploid human fibroblasts, and live cells were imaged at various times posttransfection. Whereas actin was detected on cytoplasmic filaments as well as in nuclei at 24 h, protein 4.1 initially was expressed only in nuclei, colocalizing with tagged nuclear actin (Fig. 1B). At later times, nuclear 4.1 remained coincident with actin but was also detected in the cytoplasm. Transfected cells remained alive but ceased to divide even after 72 h in culture.
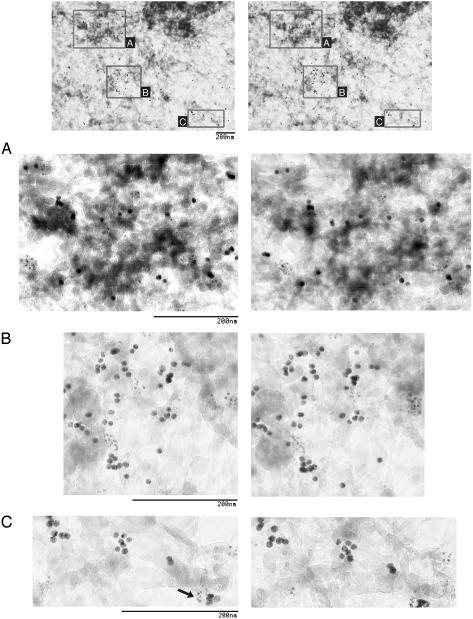
To obtain more detailed information about an association between 4.1 and actin epitopes at higher resolution, we used double-label immuno-EM. To analyze nuclear distribution of actin relative to 4.1 epitopes, we used EM of detergent-extracted cell whole mounts to visualize continuous fibrous structures not appreciated in sectioned specimens. Stereo-pair images were made of whole-mount fibroblast nuclear matrix. These were immunolabeled with different sized gold beads to detect antibodies against 4.1 SABD and actin. In these stereo pairs, clusters of beads predominately decorate fibers in areas near dense structures (Fig. 2). The 10-nm (actin epitopes) and the 5-nm (4.1 SABD epitopes) beads sometimes appear to be in subdomains of ≈100 nm or less apart on continuous fibers (Fig. 2 A and B) or sometimes intermingled (Fig. 2C, arrow, is one example). Thus the higher resolution of EM affords detailed localization of nuclear actin and 4.1 SABD epitopes in closely associated microdomains within nuclear matrix.
Fig. 2.
Immunolocalization of nuclear actin and 4.1 SABD epitopes by double-label immuno-EM of WI38 human fibroblasts. In detergent-extracted cell whole mounts, actin epitopes (10-nm beads) and 4.1 SABD epitopes (5-nm beads) decorated fibrous structures near dense structures in nuclear matrix. At the top is a stereo-pair image of a nuclear matrix region with boxes A–C indicating areas presented as stereo pairs at higher magnification in A–C, respectively. The arrow in C points to intermingled 5- and 10-nm beads.
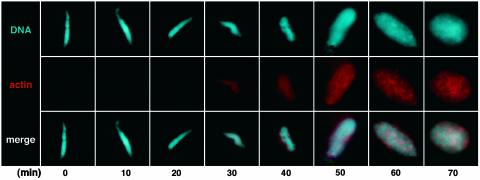
Direct Imaging of Nuclear Actin. To circumvent potential issues of fixation or overexpression artifacts, an assay was devised to image nuclear actin without using perturbing conditions. For this assay, actin covalently labeled with rhodamine was added to Xenopus egg extracts, and nuclear assembly was initiated by addition of demembranated Xenopus sperm. Nuclear actin and DNA were imaged directly in squashes of reaction aliquots by fluorescence microscopy. During the first 30 min, when sperm DNA remained highly condensed, actin was not detected in sperm pronuclei (Fig. 3). However, as DNA became decondensed and chromatin assembled, actin was detected in pronuclei (Fig. 3, 30 min). Although initially diffuse, as nuclear assembly progressed, actin accumulated and formed an apparent actin network throughout mature nuclei (Fig. 3, 60 and 70 min). In conclusion, by using a nonperturbing assay that we devised, acquisition of actin was seen to occur in concert with chromatin formation during nuclear assembly and appeared as a lattice array within mature nuclei.
Fig. 3.
Acquisition of actin during nuclear assembly in vitro. Fluorescent actin (red) was imaged as nuclei assembled in Xenopus egg extract after the addition of demembranated sperm (DNA, blue) in a representative experiment (n = 4). Diffuse actin was detected in assembling pronuclei only after 30 min. As mature nuclei assembled (50–70 min), an intranuclear actin network-like pattern became apparent.
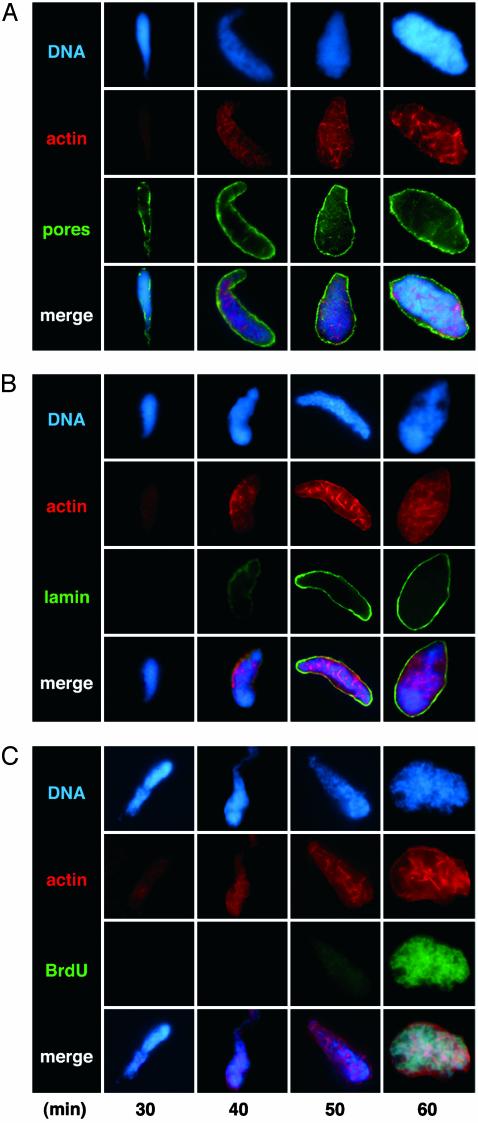
Acquisition of Nuclear Actin Relative to Nuclear Pores, Lamin, and DNA Synthesis. To determine in more detail when actin is incorporated into nuclei relative to other normal assembly markers, at various times we sedimented nuclei from reactions containing rhodamine-labeled actin onto coverslips and immunostained them in parallel for nuclear pores, the underlying lamin network, or incorporated BrdUrd in DNA. The internal rhodaminelabeled actin pattern was preserved during fixation such that secondary detection of actin was not required. Initially, when DNA was highly condensed, there was no labeling of actin, pores, lamin, or incorporated BrdUrd (data not shown). At 30 min, diffuse actin immunofluorescence was detected (as shown previously; Fig. 3) and irregularly distributed nuclear pore antigens were detected in association with chromatin (Fig. 4A). Actin continued to accumulate as pores became more regularly arrayed around chromatin. Lamin epitopes were detected at a slightly later stage than actin and pores and formed a regular rim surrounding chromatin as a distinct intranuclear actin pattern became visible (Fig. 4B). Finally, after pores and a lamina were assembled, BrdUrd incorporation was detected (Fig. 4C). Thus, nuclear actin is acquired when chromatin and nuclear pores begin to assemble and precedes lamina formation and DNA synthesis.
Fig. 4.
Acquisition of nuclear actin relative to nuclear pores, lamina formation, and DNA synthesis during nuclear assembly in vitro. Nuclei from Xenopus egg extract reactions spiked with fluorescent actin (red) were isolated at the times indicated, fixed, and probed with mAb414 against nuclear pore proteins (A, green) or antibody L046F7 against lamin (B, green). DNA synthesis was detected with anti-BrdUrd (C, green). Controls without BrdUrd had no detectable green signals. In merged images of mature nuclei, BrdUrd signals coincide with DNA (blue) as expected (aqua), and actin appears noncoincident. Diffuse actin and irregularly distributed pores are detected at 30 min, lamin epitopes at 40 min, and BrdUrd at 50 min. Assembly times vary with extract, but the relative sequence of actin acquisition, pore assembly, lamina formation, and DNA synthesis was maintained in three independent experiments.
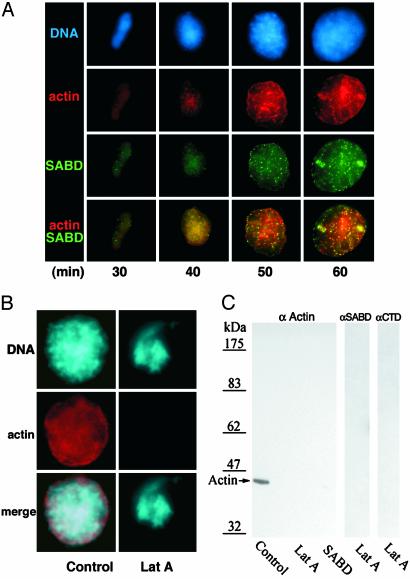
Acquisition of Nuclear Actin Relative to Protein 4.1. Because the actin-binding capacity of protein 4.1 is crucial for assembly of functional nuclei (3), we investigated incorporation of protein 4.1 into nascent nuclei relative to actin by immunostaining 4.1 SADB epitopes in nuclei isolated from in vitro reactions supplemented with fluorescent actin. SABD signals were first detected in assembling nuclei when diffuse actin signals were also detected (Fig. 5A). Initially the two signals appeared coincident (yellow coloration), but as nuclei matured actin was detected both coincident with and also independent of 4.1 epitopes. Coincident areas had strong focal staining and some diffuse staining. The larger toroidal structures (Fig. 5A, 60 min) where 4.1 and actin are coincident were shown previously to stain with antibodies against splicing factors (3).
Fig. 5.
Nuclear actin and 4.1. (A) Acquisition of actin relative to 4.1 SABD epitopes during nuclear assembly in vitro. Nuclei (DNA, blue) assembled in Xenopus egg extract spiked with fluorescent actin (red) were fixed and probed with anti-4.1 SABD (green). Data presented, from one of three independent experiments, used the same extract as described for Fig. 4. Diffuse actin and SABD epitopes detected at 30 min in the merge were largely coincident (yellow). At later times, coincidence of actin with 4.1 foci was apparent along with some areas of noncoincidence. (B) Latrunculin A (Lat A) inhibition of nuclear assembly in vitro (Right). Nuclei assembled in Xenopus egg extracts containing fluorescent actin (red) and 0.1 mM latrunculin A had highly aberrant chromatin morphology (94%; DNA, blue), and actin was not detected by fluorescence microscopy. (Left) A control nucleus. An equal concentration of DMSO (latrunculin vehicle) had no effect on nuclear assembly. (C) By Western blot analysis of equivalent numbers of nuclei from reactions with 0.1 mM latrunculin A or dominant negative 4.1 SABD peptides, actin was not detected, whereas it was readily detected in controls (Left). Nuclei perturbed by 0.1 mM latrunculin A also had no detectable 4.1 epitopes when probed with anti-4.1 SABD or anti-4.1 C terminus (αCTD) (Right).
Nuclear Actin Is Essential for Nuclear Assembly. To test whether acquisition of nuclear actin is required for nuclear assembly, the actin inhibitor latrunculin A was added to Xenopus egg extract. Latrunculin A binds to G-actin at a site adjacent to the nucleotide-binding cleft (21–23), sequestering available G-actin monomers and preventing incorporation of actin into filaments. Latrunculin A at concentrations from 1 mM down to 0.05 mM completely prevented the proper assembly of nuclei in vitro but had little effect below 10 μM (data not shown). The concentration of actin in Xenopus egg extracts has been estimated to be 15–25 μM (24). The morphologically disrupted nuclei formed in extracts containing latrunculin A were small and distorted, with areas of condensed DNA (Fig. 5B, DNA). Nuclear actin was not detected by fluorescence microscopy in perturbed nuclear structures from reactions containing rhodamine-labeled actin and latrunculin A (Fig. 5B, actin). Furthermore, defective structures had only traces of irregularly distributed pores and lamin and were incapable of DNA synthesis (data not shown).
When control nuclei assembled in vitro were analyzed by Western blotting, actin produced a strong signal but was not detected in an equal number of perturbed nuclei isolated from reactions containing latrunculin A (Fig. 5C Left), consistent with fluorescence data. Previously we showed that 4.1 SABD peptides act as dominant negatives when added to Xenopus egg extract nuclear assembly reactions, producing perturbed nuclei with no detectable 4.1 epitopes (3). When nuclei from 4.1 SABD peptide dominant negative reactions were Western-blotted, actin was not detected (Fig. 5C Left). Furthermore, protein 4.1 epitopes could not be detected by Western blotting of nuclear structures from reactions inhibited by latrunculin A (Fig. 5C Right), whereas control nuclei contain multiple 4.1 bands (3). Thus, actin acquisition is necessary for proper nuclear assembly, and inhibition of either actin or 4.1 prevents both actin and protein 4.1 from being incorporated into nuclei.
Discussion
Compelling evidence that actin is a bona fide resident nuclear protein has steadily accrued and is now widely accepted. To provide further evidence for potentially important interactions of protein 4.1 and actin in nuclei, we initially investigated the relative localization of 4.1 and actin in nuclei. We report here, using independent approaches, that 4.1 is associated with actin in nuclei at interphase. At the level of resolution afforded by detergent-extracted cell whole-mount EM, actin and 4.1 epitopes localized on continuous nuclear filaments or were even intermingled. We also devised an assay using Xenopus egg extract supplemented with fluorescent actin to directly image nuclear actin under nonperturbing conditions such that the total population of nuclear actin is retained and visualized in situ relative to intact chromatin. As best seen in the overlay of actin, DNA, and BrdUrd signals, actin appeared in mature nuclei as a filamentous array in interchromatin regions.
Recent evidence suggests that nuclear actin may be predominately monomeric G-actin, short oligomeric or branched actin polymers (refs. 25 and 26; reviewed by ref. 27). Our EM and in situ fluorescent actin data both show actin in a filamentous pattern but do not address whether any nuclear actin is F-actin or whether its distribution reflects association with a filamentous nuclear structure or a channel-like space. Although in Drosophila the EAST protein controls an expandable nuclear endoskeleton that promotes actin accumulation in extrachromosomal regions (28), filament proteins in vertebrate nuclei are not well characterized. When these are identified, we may better understand their relationship to nuclear actin.
Cell-free Xenopus egg extracts are a powerful in vitro experimental system that recapitulates in vivo events of nuclear assembly and DNA replication. Nuclear reconstitution with Xenopus egg extracts and demembranated sperm has been characterized as proceeding through discrete intermediate steps (29, 30): DNA forms nucleosomes and scaffold proteins bind and organize chromatin; vesicles and pores bind to chromatin; lamins polymerize to form a lamina; membrane vesicles fuse to form a double membrane envelop; and chromatin decondenses and DNA replication centers form as nuclei further mature.
We first detected diffuse nuclear actin during initial stages of chromatin assembly in egg extracts. Actin continued to accumulate in nuclei during assembly and became organized into a discrete network during lamina and pore formation before DNA synthesis. Therefore, acquisition of nuclear actin seems to be an early event. This observation is supported by our finding that latrunculin A inhibition of nuclear assembly resulted in small irregular nuclear structures often with highly condensed DNA. These morphologically abnormal structures had only traces of detectable pores and lamin and were defective in synthesizing DNA, suggesting that nuclear actin may even be a prerequisite for organization of functional pores, lamina, and DNA replication centers. In a previous preliminary report also with Xenopus egg extracts, the actin polymerization inhibitor gelsolin S1 inhibited nuclear formation and DNA replication.¶ The latrunculin A nuclear phenotype we observe is quite similar to that resulting from 4.1 depletion or by dominant negative 4.1 peptides (3) and distinct from the effects of lamin depletion (31) or inhibition of nuclear pores (32), again suggesting that inhibition of actin acquisition interferes with nuclear assembly at a very early stage.
The function of actin in the nucleus is an area under intense investigation. Actin has been shown to be integral to a number of chromatin-modifying complexes having ATPase, helicase, and acetylase activities (10). Actin has also been reported to complex with members of the A/B-group heterogeneous nuclear ribonucleoprotein (hnRNP) proteins (33). A nuclear complex was identified containing nuclear DNA helicase II (NDH II), heterogeneous nuclear ribonucleoprotein C (hnRNP C), and actin (34). Multiple lines of evidence implicate actin in mRNA processing and export: Rapidly labeled RNAs associate with actin in nuclear matrix (35), cytochalasin B causes selective release of precursor mRNA (36), actin localizes adjacent to snRNP aggregates (37), and actin associates with intron-containing HIV-1 gag mRNA (38) and is essential for revmediated mRNA export (39).
What roles might 4.1 play in the nucleus in conjunction with actin? Our data show that actin and 4.1 both become diffusely associated with nascent chromatin in early phases of nuclear assembly. Actin contains two nuclear export signals but no nuclear localization signal, and thus it is unclear how actin becomes nuclear. Several 4.1 family members have actin-binding domains, and 4.1R has sites regulating nuclear localization distinct from its actin-binding site (40–42). It is possible that one regulated means for actin to become nuclear is via 4.1 binding, a hypothesis to be tested in the future. At later stages of nuclear assembly, 4.1 remains extensively colocalized with actin in foci, although actin also forms an extensive intranuclear network. Some areas of 4.1–actin colocalization may be mRNA-splicing centers, because 4.1 colocalizes with splicing centers in mammalian cells (1, 43) and in Xenopus nuclei assembled in vitro (3), 4.1 coimmunoprecipitates with splicing factors, and 4.1 depletion inhibits RNA splicing in vitro (44). Other colocalization signals may detect complexes of 4.1, actin, and actin-binding proteins. For example, we have preliminary evidence that 4.1 can bind the BAF (BRG and Brm associated factors) chromatin remodeling complex (S.W.K., unpublished observations). Additionally, we found that incubation of mature nuclei with latrunculin A causes nuclear structures to become distorted and chromatin to collapse (S.W.K., unpublished observations), which suggests that nuclear actin is essential for maintenance of interphasic nuclear structure as well as for nuclear assembly. It is also interesting that mammalian cells cease dividing when actin and 4.1 are overexpressed by cotransfection or overexpressed individually (S.W.K., unpublished observations). Perhaps relative levels of nuclear 4.1 and actin are highly regulated during normal cell cycling.
Previously we showed that an intact actin-binding site in protein 4.1 is essential for nuclear assembly, and here we report that sequestration of actin monomers with latrunculin A both prevented 4.1 incorporation into nuclei and inhibited nuclear assembly. It will be important to test whether latrunculin A directly prevents actin–4.1 SABD binding. Protein 4.1 interactions with actin in nuclei may recruit, target, and/or modulate functions of various actin-containing complexes, a role reminiscent of its well established adaptor function in the erythrocyte membrane skeleton.
Acknowledgments
S.W.K. dedicates this paper to the memory of Professor Morris E. Friedkin. We thank G. Krockmalnic for expertise in detergent-extracted cell whole-mount EM; Drs. N. Mohandas, D. Drubin, M. Welch, R. Jeng, and D. Nicks for helpful discussions; and R. Couto for aid in figure preparation. This research was supported by National Institutes of Health Grants DK59079 (to S.W.K.) and GM57839 (to R.H.).
Abbreviations: SABD, spectrin–actin binding domain; EM, electron microscopy; YFP, yellow fluorescent protein; CFP, cyan fluorescent protein.
Footnotes
Zhang, C., Way, M. & Hutchison, C. J. (1996) Mol. Biol. Cell 7, 477 (abstr.).
References
- 1.Krauss, S. W., Larabell, C. A., Lockett, S., Gascard, P., Mohandas, N. & Chasis, J. A. (1997) J. Cell Biol. 137, 275–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Krauss, S. W., Chasis, J. A., Rogers, C., Mohandas, N., Krockmalnic, G. & Penman, S. (1997) Proc. Natl. Acad. Sci. USA 94, 7297–7302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Krauss, S. W., Heald, R., Lee, G., Nunomura, W., Gimm, J. A., Mohandas, N. & Chasis, J. A. (2002) J. Biol. Chem. 277, 44339–44346. [DOI] [PubMed] [Google Scholar]
- 4.Peters, L. L., Weier, H. U., Walensky, L. D., Snyder, S. H., Parra, M., Mohandas, N. & Conboy, J. G. (1998) Genomics 54, 348–350. [DOI] [PubMed] [Google Scholar]
- 5.Conboy, J. G. (1993) Semin. Hematol. 30, 58–73. [PubMed] [Google Scholar]
- 6.Benz, E. J., Jr. (1994) in The Molecular Basis of Blood Diseases, eds. Stamatoyannopoulos, G., Neinhuis, A. W., Majerus, P. & Varmus, H. (Saunders, Philadelphia), pp. 257–292.
- 7.Clark, T. G. & Merriam, R. W. (1977) Cell 12, 883–891. [DOI] [PubMed] [Google Scholar]
- 8.Capco, D. G., Wan, K. M. & Penman, S. (1982) Cell 29, 847–858. [DOI] [PubMed] [Google Scholar]
- 9.Nakayasu, H. & Ueda, K. (1985) Cell Struct. Funct. 10, 305–309. [DOI] [PubMed] [Google Scholar]
- 10.Olave, I. A., Reck-Peterson, S. L. & Crabtree, G. R. (2002) Annu. Rev. Biochem. 71, 755–781. [DOI] [PubMed] [Google Scholar]
- 11.Wada, A., Fukuda, M., Mishima, M. & Nishida, E. (1998) EMBO J. 17, 1635–1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rando, O. J., Zhao, K. & Crabtree, G. R. (2000) Trends Cell Biol. 10, 92–97. [DOI] [PubMed] [Google Scholar]
- 13.Pestic-Dragovich, L., Stojiljkovic, L., Philimonenko, A. A., Nowak, G., Ke, Y., Settlage, R. E., Shabanowitz, J., Hunt, D. F., Hozak, P. & de Lanerolle, P. (2000) Science 290, 337–341. [DOI] [PubMed] [Google Scholar]
- 14.Tse, W. T., Tang, J., Jin, O., Korsgren, C., John, K. M., Kung, A. L., Gwynn, B., Peters, L. L. & Lux, S. E. (2001) J. Biol. Chem. 276, 23974–23985. [DOI] [PubMed] [Google Scholar]
- 15.Pendleton, A., Pope, B., Weeds, A. & Koffer, A. (2003) J. Biol. Chem. 278, 14394–14400. [DOI] [PubMed] [Google Scholar]
- 16.Murray, A. (1991) Methods Cell Biol. 36, 581–605. [PubMed] [Google Scholar]
- 17.Pockwinse, S. M., Krockmalnic, G., Doxsey, S. J., Nickerson, J., Lian, J. B., van Wijnen, A. J., Stein, J. L., Stein, G. S. & Penman, S. (1997) Proc. Natl. Acad. Sci. USA 94, 3022–3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Verheijen, R., Kuijpers, H., Vooijs, P., van Venrooij, W. & Ramaekers, F. (1986) J. Cell Sci. 80, 103–122. [DOI] [PubMed] [Google Scholar]
- 19.De Carcer, G., Lallena, M. J. & Correas, I. (1995) Biochem. J. 312, 871–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peters, K. E., Okada, T. A. & Comings, D. E. (1982) Eur. J. Biochem. 129, 221–232. [DOI] [PubMed] [Google Scholar]
- 21.Belmont, L. D., Patterson, G. M. & Drubin, D. G. (1999) J. Cell Sci. 112, 1325–1336. [DOI] [PubMed] [Google Scholar]
- 22.Morton, W. M., Ayscough, K. R. & McLaughlin, P. J. (2000) Nat. Cell Biol. 2, 376–378. [DOI] [PubMed] [Google Scholar]
- 23.Yarmola, E. G., Somasundaram, T., Boring, T. A., Spector, I. & Bubb, M. R. (2000) J. Biol. Chem. 275, 28120–28127. [DOI] [PubMed] [Google Scholar]
- 24.Rosenblatt, J., Peluso, P. & Mitchison, T. J. (1995) Mol. Biol. Cell 6, 227–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhao, K., Wang, W., Rando, O. J., Xue, Y., Swiderek, K., Kuo, A. & Crabtree, G. R. (1998) Cell 95, 625–636. [DOI] [PubMed] [Google Scholar]
- 26.Gonsior, S. M., Platz, S., Buchmeier, S., Scheer, U., Jockusch, B. M. & Hinssen, H. (1999) J. Cell Sci. 112, 797–809. [DOI] [PubMed] [Google Scholar]
- 27.Pederson, T. & Aebi, U. (2002) J. Struct. Biol. 140, 3–9. [DOI] [PubMed] [Google Scholar]
- 28.Wasser, M. & Chia, W. (2000) Nat. Cell Biol. 2, 268–275. [DOI] [PubMed] [Google Scholar]
- 29.Newport, J. (1987) Cell 48, 205–217. [DOI] [PubMed] [Google Scholar]
- 30.Almouzni, G. & Wolffe, A. P. (1993) Exp. Cell Res. 205, 1–15. [DOI] [PubMed] [Google Scholar]
- 31.Jenkins, H., Holman, T., Lyon, C., Lane, B., Stick, R. & Hutchison, C. (1993) J. Cell Sci. 106, 275–285. [DOI] [PubMed] [Google Scholar]
- 32.Macaulay, C. & Forbes, D. J. (1996) J. Cell Biol. 132, 5–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Percipalle, P., Zhao, J., Pope, B., Weeds, A., Lindberg, U. & Daneholt, B. (2001) J. Cell Biol. 153, 229–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang, S., Buder, K., Burkhardt, C., Schlott, B., Gorlach, M. & Grosse, F. (2002) J. Biol. Chem. 277, 843–853. [DOI] [PubMed] [Google Scholar]
- 35.Nakayasu, H. & Ueda, K. (1985) Exp. Cell Res. 160, 319–330. [DOI] [PubMed] [Google Scholar]
- 36.Schroder, H. C., Trolltsch, D., Wenger, R., Bachman, M., Diehl-Seifert, B. & Muller, W. E. G. (1987) Eur. J. Biochem. 167, 239–245. [DOI] [PubMed] [Google Scholar]
- 37.Sahlas, D. J., Milankov, K., Park, P. C. & De Boni, U. (1993) J. Cell Sci. 105, 347–357. [DOI] [PubMed] [Google Scholar]
- 38.Kimura, T., Hashimoto, I., Yamamoto, A., Nishikawa, M. & Fujisawa, J. I. (2000) Genes Cells 5, 289–307. [DOI] [PubMed] [Google Scholar]
- 39.Hofmann, W., Reichart, B., Ewald, A., Muller, E., Schmitt, I., Stauber, R. H., Lottspeich, F., Jockusch, B. M., Scheer, U., Hauber, J. & Dabauvalle, M. C. (2001) J. Cell Biol. 152, 895–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gimm, J. A., An, X., Nunomura, W. & Mohandas, N. (2002) Biochemistry 41, 7275–7282. [DOI] [PubMed] [Google Scholar]
- 41.Gascard, P., Nunomura, W., Lee, G., Walensky, L., Krauss, S., Chasis, J., Mohandas, N. & Conboy, J. (1999) Mol. Biol. Cell 10, 1783–1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Luque, C. M., Perez-Ferreiro, C. M., Perez-Gonzalez, A., Englmeier, L., Koffa, M. D. & Correas, I. (2003) J. Biol. Chem. 278, 2686–2691. [DOI] [PubMed] [Google Scholar]
- 43.Lallena, M. J. & Correas, I. (1997) J. Cell Sci. 110, 239–247. [DOI] [PubMed] [Google Scholar]
- 44.Lallena, M. J., Martinez, C., Valcarcel, J. & Correas, I. (1998) J. Cell Sci. 111, 1963–1971. [DOI] [PubMed] [Google Scholar]