Abstract
Different components of the AP1 transcription factor complex appear to have distinct effects on cell proliferation and transformation. In contrast to other AP1 components, JunD has been shown to inhibit cell proliferation. Also, in prior studies, JunD alone bound menin, product of the MEN1 tumor suppressor gene, and JunD's transcriptional activity was inhibited by menin, suggesting that JunD might achieve all or most of its unique properties through binding to menin. Analyses of JunD and menin effects on proliferation, morphology, and cyclin D1 in stable cell lines unmasked an unexpected growth promoting activity of JunD. Whereas stable overexpression of wild-type (wt) mouse JunD in JunD–/– immortalized fibroblasts inhibited their proliferation and reverted their transformed-like phenotype, overexpression of a missense mouse JunD mutant (mJunDG42E) with disabled binding to menin showed opposite or growth promoting effects. Similarly, stable overexpression of wt mouse JunD in wt immortalized fibroblasts inhibited growth. In contrast, its overexpression in Men1–/– immortalized fibroblasts enhanced their already transformed-like characteristics. To conclude, JunD changed from growth suppressor to growth promoter when its binding to menin was prevented by a JunD mutant unable to bind menin or by Men1-null genetic background.
JunD belongs to the Jun family of the AP1 transcription factor complex. The Jun proteins via their basic-leucine zipper (bZIP) domains can form homodimers or form heterodimers with the bZIP domains of Fos and the activating transcription factor/cAMP-response element-binding protein (ATF/CREB) proteins; these dimers bind to promoters at specific DNA elements (TRE or CRE sites) and regulate transcription (1). These AP1 proteins are important in tumorigenesis and other processes (2, 3), so JunD, like other AP1 proteins, could also contribute to neoplasia. JunD binds directly to menin, which is encoded by the MEN1 tumor suppressor gene (4, 5). In fact, among all of the AP1 transcription factors, only JunD binds directly to menin (4).
MEN1 mutation causes hereditary and nonhereditary tumors (6, 7). The tumor suppressor function of menin is supported (i) by germ-line and somatic mutations of MEN1 that mostly predict menin protein truncation or absence, and (ii) by accompanying tumor-specific loss of chromosomal alleles about the MEN1 locus, which is consistent with biallelic menin inactivation (7). In addition, overexpression of menin in Ras-transformed NIH3T3 cells partially reverts the transformed phenotype, further supporting menin's growth suppressor function (8). However, the mechanism by which menin might act as a growth suppressor in concert with JunD or with any of its other 10 identified protein partners is unknown (9, 10).
JunD and menin interact near the N terminus of each. Engineered JunD missense mutations at amino acids 41–44 in the menin-binding region of mouse JunD, and in a conserved amino acid in the corresponding region of human JunD (amino acid 34), disrupt mutant JunD binding to menin, and also disrupt the ability of menin to suppress the transcriptional activity of the mutant JunD (11); effects, if any, of these JunD missense mutations on growth have not been reported. In a reciprocal manner, several tumorigenic, missense mutations in menin (mostly in the region from amino acids 139–242) disrupt its binding to JunD, and also disrupt the ability of mutant menin to repress JunD transcriptional activity (4).
JunD behaves differently from related cJun and JunB proteins (12). Overexpression of cJun, and to a lesser extent, JunB, is capable of transforming avian and rodent fibroblasts (13). Furthermore, cJun can also cooperate with Ras to transform rodent fibroblasts (12). In contrast, overexpression of JunD in rodent fibroblasts suppresses proliferation and also antagonizes Ras-mediated transformation (12). Furthermore, fibroblasts from cJun–/– mice exhibit a defect in proliferation even after immortalization (14). In contrast, immortalized JunD–/– mouse embryo fibroblasts show increased proliferation (15). Given the growth suppressor properties of JunD, it would be paradoxical for menin's growth suppressor function to be mediated mainly by inhibiting the activity of JunD, another growth suppressor. Herein, growth indices are explored for JunD with or without compromise of its possibility to bind menin.
Materials and Methods
Cell Culture, Transfection, and Cyclin D1 Assay. Cell lines were grown in complete DMEM supplemented with 10% FCS, 2 μM glutamine, and 100 μg of penicillin–streptomycin per ml at 5% CO2. Transfections were carried out with Superfect reagent (Qiagen, Valencia, CA).
Cells were synchronized for cyclin D1 testing (16). Immortalized fibroblasts were plated in triplicate, and one plate was used for making whole-cell extracts of exponentially growing cells (E). Two plates were serum-starved by culturing in medium with 0.5% FCS for 48 h; after 48 h, one of the plates was used to make extracts of serum-starved cells (SS). The other plate was cultured in medium with 20% FCS for another 5–6 h to generate extracts of serum-induced cells (SI).
Immortalized Fibroblasts. JunD-HET-1 and JunD-HET-2 (Yaniv Laboratory names HT1 and HT2) are JunD–/– spontaneously immortalized fibroblasts from embryonic day (E)12.5–E14 heterozygote (HET) JunD–/– mouse embryos in which one copy of the entire JunD coding sequence has been replaced with a bacterial NLS-LacZ cassette (15, 17). JunD-NULL-1 and JunD-NULL-2 (Yaniv Laboratory names HM1 and HM2) are JunD–/– spontaneously immortalized fibroblasts from E12.5–E14 mouse embryos (17). JunD-WT (Yaniv Laboratory name WT) is a spontaneously immortalized fibroblast line matched for the JunD-HET and JunD-NULL genetic background.
Multiple endocrine neoplasia type 1 (Men1)-NULL-17 and Men1-NULL-41 are Men1–/– spontaneously immortalized fibroblasts from E8.5–E10.5 mouse embryos in which Men1 exons 3–8 was homozygously deleted (18). Men1-WT-10 is a line of wt immortalized fibroblasts of this same preparation and genetic background, which is a different genetic background than for JunD-WT. Men1 wt and Men1–/– immortalized fibroblasts will be described elsewhere.
Stable Cell Lines. Plasmid constructs for stable transfections were made in the pcDNA3.1-hygro vector (Invitrogen). The inserts from pcDNA3.1-mouse-JunD, pcDNA3.1-mouse-JunD G42E, or pcDNA3.1-cJun (4, 11) were excised with HindIII/NotI and ligated into the corresponding sites of the pcDNA3.1-hygro vector (named pcDNA3.1-hygro-mJunD, pcDNA3.1-hygro-mJunDG42E, and pcDNA3.1-hygro-cJun). Immortalized fibroblasts were transfected and selected in medium containing hygromycin-B, 300 μg/ml for Men1-WT-10 and Men1-NULL-17; 500 μg/ml for JunD-NULL-2; and 600 μg/ml for Men1-NULL-41. After 2 weeks of selection, independent hygromycin-resistant colonies were collected individually with cloning cylinders and then expanded. Names of stably transfected cell lines were suffixed as follows: vector (V), mJunD (JD), mJunDG42E (G42E), and cJun (JC). The amount of transfected protein in each line was determined for several passages by Western blot and immunofluorescence. Cell lines for comparison were matched on the same genetic background and passage. Microscopy and photomicrography were performed with an Axiovert-450 inverted phase contrast microscope (Zeiss).
Western Blots. For Western blot of nuclear extracts (NE), immortalized fibroblasts were cultured in 10-cm plates and washed with 1× Dulbecco's PBS (DPBS); NE were prepared by using the NEPER kit (Pierce). For whole-cell extracts (WCE), cells were washed with 1× DPBS and lysed in 1× lysis buffer (Promega). Protein samples (25 μg for NE and 50 μg for WCE) were separated in 10% Tris-glycine SDS/PAGE gels (Invitrogen), transferred to nitrocellulose membranes, and hybridized with appropriate antibodies [SQV (19); JunD (sc-74), cJun (sc-1694), and cyclin D1 (sc-718) from Santa Cruz Biotechnology]. Blots were developed by enhanced chemiluminescence. Antibodies against p84 (Genetex) or NUMA (Babco, Richmond, CA) were used to compare membrane loading.
Proliferation Assay. Proliferation was tested by a colorimetric assay using the CellTiter 96 aqueous one solution cell proliferation assay kit (Promega). Cells (1,500–3,000 per well) were plated in triplicate in 96-well flat-bottomed plates in 100 μl of complete DMEM or complete DMEM with the appropriate amount of hygromycin B. Proliferation was monitored later in the day (day 1) and/or every other day (days 2, 4, 6, and 8 after plating). For proliferation in low serum, 24 h after the cells were plated, the medium was changed to 100 μl of DMEM containing 0.5% FCS supplemented with the appropriate amount of hygromycin-B. All proliferation assays were performed as at least three independent experiments at separate times.
Colony Formation in Soft Agar. Assays were performed in duplicate (8). In brief, 5,000–6,000 cells were resuspended in 3 ml of molten 0.33% agar and poured over a 5-ml solidified layer of 0.66% agar made in complete DMEM. Agar plates were incubated for 2 weeks and observed for colony formation.
JunD Mutation Analysis. DNA from non-MEN1 parathyroid tumors (20), insulinomas, and gastrinomas was analyzed. The N-terminal 100-aa region of hJunD was amplified from tumor DNA and two control DNA samples by PCR, using primers hJD1A-GCCGAATTCCGGAGGATGGAAACACCCTTCTAC and hJD1C-GCCGGATCCCTTCAGCAGCCCCAGGTCGGGAGAG, with TaqGold (Applied Biosystems). PCR products were sequenced with primer JD1-CGGAGGATGGAAACACCCTTCTAC by using an ABI3700 automated sequencer (MWG Biotech, High Point, NC).
Results
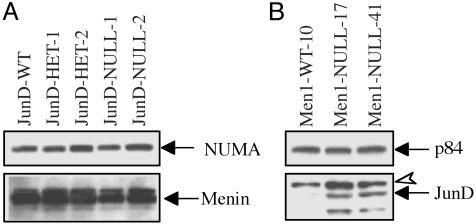
JunD or Menin as Possible Stable Regulators of Each Other's Concentration. For analysis of effects of JunD on menin expression, Western blots were performed by using nuclear extracts from immortalized fibroblasts that were JunD–/– (JunD-WT), JunD–/– (JunD-HET-1 and JunD-HET-2) or JunD–/– (JunD-NULL-1 and JunD-NULL-2). Relative expression of menin was similar in all cell lines (Fig. 1A).
Fig. 1.
Menin expression in JunD–/– immortalized fibroblasts and JunD expression in Men1–/– immortalized fibroblasts. (A) Western blot of nuclear extracts (25 μg) from immortalized fibroblasts, JunD-WT, JunD-heterozygote, or JunD-null. Blot was probed with an anti-menin antibody (SQV) (Lower) and reprobed with anti-NUMA antibody as loading control and nuclear protein marker (Upper). Lower arrow points to the menin-specific band. Decrease or absence of JunD did not affect menin expression. (B) Western blot of nuclear extracts (25 μg) from immortalized fibroblasts, which are wt or Men1-null. Blot was probed with an anti-JunD antibody (Lower) and reprobed with the p84 antibody as loading control and nuclear protein marker (Upper). Solid black arrow below indicates the JunD-specific band, and the open arrowhead denotes a nonspecific band. Both Men1–/– lines express far more JunD than wt. The faster migrating band below the JunD likely represents an alternate smaller form of JunD generated from an internal translation initiation site (22).
Furthermore, nuclear extracts were evaluated from immortalized fibroblasts that were either Men1–/– (Men1-WT-10) or Men1–/– (Men1-NULL-17 and Men1-NULL-41). Both Men1–/– immortalized fibroblast lines expressed far higher levels of JunD (Fig. 1B). This finding suggests a regulatory role for menin on JunD expression and possibly a contribution from increased JunD to the menin-null phenotype.
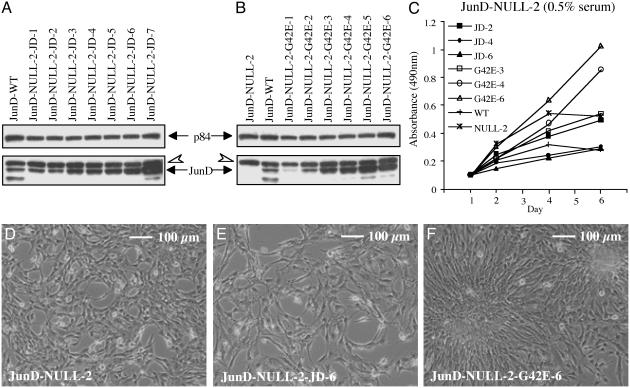
Stable Overexpression of mJunD or mJunDG42E in JunD-Null Cells: Proliferation and Morphology. JunD–/– immortalized fibroblasts (JunD-NULL-2) with normal menin levels (above) were stably transfected with vector alone, mJunD, or mJunDG42E (mJunD missense mutant that cannot bind to menin). Three independent stable lines each of vector as control or overexpressing either mJunD or mJunDG42E with matched expression levels were studied (Fig. 2 A and B).
Fig. 2.
mJunD and mJunD mutant protein expression, proliferation, and morphology of JunD–/– stable lines. Cell morphology under phase contrast was examined in 10% FCS, and proliferation was analyzed in 0.5% FCS. (A) Western blot of nuclear extracts (25 μg) from JunD-null immortalized fibroblasts stably transfected with mJunD (from JunD-NULL-2-JD-1 to -JD7). (B) Western blot of nuclear extracts (25 μg) from JunD-null immortalized fibroblasts stably transfected with mutant mJunDG42E (from JunD-NULL-2-G42E-1 to -G42E-6). Blots in A and B were probed with an anti-JunD antibody (Lower) and reprobed with the p84 antibody as loading control (Upper). Untransfected JunD-null and wt is also shown as control. The open arrowhead shows a nonspecific band also seen in the JunD-null cells. The solid arrow shows full-length JunD. The faster migrating band below the JunD band likely represents an alternate smaller form of JunD generated from an internal translation initiation site (22). (C) Proliferation curves for three independent JunD-NULL-2 stable lines overexpressing mJunD or overexpressing mutant mJunDG42E that express numerically increasing protein from transfection. Proliferation curves for JunD-NULL-2 and a genetically matched wt line are shown for comparison. Three independent lines of JunD-NULL-2 stably transfected with vector alone showed proliferation curves similar to JunD-NULL-2 (data not shown). Each point represents the mean of triplicate cultures from the same experiment. Standard deviations were 5–20% of the mean (data not shown). (D) Morphology of JunD–/– immortalized fibroblasts. Cells are spindle-like and pile up at high cell density. (E) Morphology of JunD–/– cells stably overexpressing mJunD. Compared with JunD–/–, cells are flattened and are highly spread. (F) Morphology of JunD–/– cells stably overexpressing mutant mJunDG42E. Cells pile up to form foci at high cell densities.
As previously reported (15), the proliferation rate of JunD-NULL-2 was increased, compared with JunD-WT (Fig. 2C). In medium containing low serum (0.5% FCS) and compared with JunD-NULL-2 or vector-transfected JunD-NULL-2 (data not shown), JunD-NULL-2-JD-4 and JunD-NULL-2-JD-6, expressing higher levels of mJunD than wt, showed reduced proliferation, whereas JunD-NULL-2-G42E-4 and JunD-NULL-2-G42E-6 expressing higher levels of mJunDG42E showed increased proliferation (Fig. 2C). In medium containing 10% FCS, untransfected or stably transfected JunD-NULL-2 (vector or JunDG42E) displayed some lack of cell adhesion at confluency, and detached from the culture plate on or after day 4 of analysis; consequently, these growth curves were moderately variable, but the comparisons (data not shown) were similar to those in 0.5% FCS.
The terms “transformed-like” or “transforming-like” are applied herein to fibroblasts with some if not all of the following: accelerated proliferation, ability to proliferate in 0.5% FCS, spindle-shaped refractile morphology, lack of contact inhibition (cells pile up), focus formation, and colony formation in soft agar (21). These terms imply that proliferation rate or morphology alone cannot be used for prediction of all aspects of transformed behavior (21).
As reported previously (15), JunD–/– immortalized fibroblasts showed some transformed-like features, specifically, a spindle-like morphology, and a tendency to pile up at high-cell density (Fig. 2D). In the current analysis they also formed small colonies in soft agar (data not shown). mJunD overexpression in JunD–/– immortalized fibroblasts (JunD-NULL-2-JD-4 and JunD-NULL-2-JD-6) resulted in a different morphology with flattened highly spread cells (Fig. 2E), which did not pile up and did not form colonies in soft agar. In contrast, mJunDG42E overexpressing lines (JunD-NULL-2-G42E-4 and JunD-NULL-2-G42E-6) differed from the parental JunD-NULL-2 in that they formed foci (Fig. 2F), and they were similar to JunD-NULL-2 in forming small colonies in soft agar.
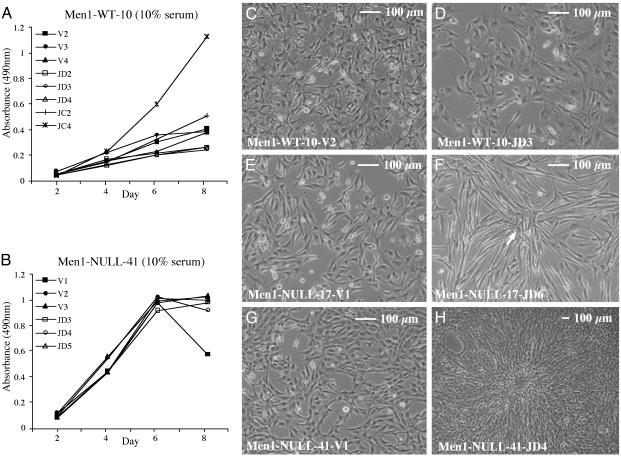
Stable Overexpression of mJunD in Men1-Null Cells: Proliferation and Morphology. Given the opposite effects of mJunD and mJunDG42E on proliferation and morphology in JunD–/– immortalized fibroblasts (above), it seemed possible that the negative effect of JunD on growth indices depended on its binding to menin. To test this hypothesis further, vector or mJunD were stably transfected into Men1–/– immortalized fibroblasts (Men1-WT-10) and into two independent Men1–/– immortalized fibroblasts (Men1-NULL-17 and Men1-NULL-41). cJun was transfected into Men1-WT-10 for comparison. Levels of the transfected Jun proteins are shown (see Fig. 5, which is published as supporting information on the PNAS web site, www.pnas.org).
In low serum (0.5% FCS), the proliferation of Men1-WT-10 cells transfected with vector or mJunD was slow, but was faster in the cJun-transfected line Men1-WT-10-JC4 (with highest cJun expression; data not shown). In 10% FCS, the proliferation of all three mJunD-transfected lines (Men1-WT-10-JD2, Men1-WT-10-JD3, and Men1-WT-10-JD4) was slower than vector-transfected controls, but faster in Men1-WT-10-JC4 (Fig. 3A). In both 0.5% (data not shown) and 10% FCS, mJunD transfection into Men1-NULL-17 and Men1-NULL-41 caused no inhibition or increase of proliferation (Fig. 3B; Men1-NULL-17; data not shown), and contrasted with the inhibition observed in Men1-WT-10 (Fig. 3A).
Fig. 3.
Proliferation and morphology of stable lines based on wt immortalized fibroblasts or Men1–/– immortalized fibroblasts. Cell morphology under phase contrast or proliferation were examined in 10% FCS. Each point in the growth curve represents the mean of triplicate cultures from the same experiment. Standard deviations were 3–15% of the mean (data not shown). (A) Men1-WT-10 transfected with mJunD and two independent lines transfected with cJun that express numerically increasing protein from transfection. Lines stably transfected with vector are shown for comparison. (B) Men1-NULL-41 vector transfected lines and three independent mJunD-transfected lines that express numerically increasing levels of mJunD. The proliferation of mJunD expressing Men1-NULL-41 was unaltered compared with vector control. Men1-NULL-17 gave similar results (data not shown). (C) Vector-transfected line. (D) mJunD-transfected line. Cells are more flattened and more highly spread than in C.(E) Vector-transfected line Men1-NULL-17-V1. Cells are spindle-shaped, compared with vector-transfected Men1-WT-10 (A). (F) mJunD-transfected line Men1-NULL-17-JD6. Cells are refractile, are elongated, and pile up. (G) Vector-transfected line Men1-NULL-41-V1. Cells are spindle-shaped, compared with vector-transfected Men1-WT-10 (C). The cells pile up at high cell densities (data not shown). (H) mJunD-transfected line Men1-NULL-41-JD4. Cells are refractile, slightly elongated, and pile up to form foci (one focus occupies the center of the image).
Compared with vector-transfected Men1-WT-10 (Fig. 3C), Men1-WT-10 overexpressing mJunD showed a more flattened and highly spread morphology (Fig. 3D); in contrast, cJun overexpressing cells were highly refractile and piled up to form foci (see Fig. 6A, which is published as supporting information on the PNAS web site). Consistent with previous reports for NIH3T3 fibroblasts, vector-transfected Men1-WT-10 or Men1-WT-10 overexpressing mJunD did not form colonies in soft agar, and Men1-WT-10 overexpressing cJun (Men1-WT-10-JC4) formed small colonies in soft agar (Fig. 6B and refs. 12 and 22).
Men1-NULL-17 (Fig. 3E) stably overexpressing mJunD was highly elongated and had a highly refractile morphology, piled up, but did not form foci (Fig. 3F) or colonies in soft agar (data not shown). Men1-NULL-41 (Fig. 3G) stably overexpressing mJunD was slightly elongated, piled up to form foci (Fig. 3H), and formed colonies in soft agar that were larger than the colonies of the parental Men1-NULL-41 (see Fig. 7, which is published as supporting information on the PNAS web site). The more modest effect on transformed-like morphology from JunD overexpression in Men1-NULL-17, as compared with that seen in JunD overexpressing Men1-NULL-41, could be due to the difference in the expression level of the transfected mJunD (Fig. 5), or due to the presence of some greater inherent transformed-like characteristics in Men1-NULL-41. Overall, mJunD overexpression caused both Men1-NULL-17 and Men1-NULL-41 to show shifts in the same direction; i.e., toward a more transformed-like morphology.
Cyclin D1 Expression. Overexpression of JunD in immortalized fibroblasts arrests cells at G0-G1 (12) and increasing the JunD:cJun ratio decreases cyclin D1 expression (23). Cyclin D1 levels were investigated alongside other growth-related indices. Cyclin D1 protein levels were measured in cells grown exponentially, serum deprived, or grown in medium with 20% FCS after synchronization by serum deprivation. The results are shown in Table 1, and Fig. 8, which is published as supporting information on the PNAS web site.
Table 1. Cyclin D1 protein level in untransfected or stably transfected immortalized fibroblasts.
| E
|
ST
|
SI
|
|
|---|---|---|---|
| Immortalized fibroblasts | Cyclin D1 protein* | ||
| JunD-WTa | + | 0 | ++ |
| JunD-NULL-2a | ++ | + | +++ |
| JunD-NULL-2-JD4a | ++ | 0 | 0 |
| JunD-NULL-2-JD7a | ++ | 0 | + |
| JunD-NULL-2-G42E-4a | ++++ | ++++ | ++++ |
| JunD-NULL-2-G42E-6a | ++++ | ++++ | ++++ |
| Men1-WT-10-V1bcd | + | + | ++ |
| Men1-WT-10-V2bcd | + | 0 | + |
| Men1-WT-10-JD4b | + | 0 | 0 |
| Men1-WT-10-JC4b | ++++ | +++ | ++++ |
| Men1-NULL-17-V1c | +++ | + | +++ |
| Men1-NULL-17-V2c | ++ | 0 | +++ |
| Men1-NULL-17-V3c | ++ | 0 | +++ |
| Men1-NULL-17-JD4c | ++ | 0 | ++ |
| Men1-NULL-17-JD5c | ++ | 0 | ++ |
| Men1-NULL-17-JD6c | +++ | 0 | +++ |
| Men1-NULL-41-V1d | ++ | ++ | ++ |
| Men1-NULL-41-V2d | ++ | ++ | ++ |
| Men1-NULL-41-V3d | +++ | +++ | +++ |
| Men1-NULL-41-JD3d | ++ | ++ | ++ |
| Men1-NULL-41-JD4d | ++ | ++ | ++ |
| Men1-NULL-41-JD5d | +++ | +++ | +++ |
Cyclin D1 level determined by Western blot of cells growing exponentially (E), serum-starved (ST), or serum-induced (SI). Superior letters represent groups compared within an experiment. +, cyclin D1 level (+, lowest; ++++, highest); 0 not detectable.
As expected, cyclin D1 protein expression in JunD-WT cells decreased after serum deprivation and increased after serum addition. In agreement with previous results (15), JunD–/– immortalized fibroblasts expressed higher cyclin D1 levels than did wt immortalized fibroblasts. In JunD–/– immortalized fibroblasts overexpressing mJunD, serum stimulation failed to increase cyclin D1 (a result similar to wt immortalized fibroblasts overexpressing mJunD). However, with overexpression of missense mutant mJunDG42E in JunD–/– immortalized fibroblasts, cyclin D1 level was higher under all conditions.
Men1-WT-10 vector controls, like JunD-WT, showed decrease of cyclin D1 after serum starvation and increase of cyclin D1 after serum stimulation. Again, as expected, Men1-WT-10 overexpressing mJunD (Men1-WT-10-JD4) failed to increase cyclin D1 after serum stimulation. However, the cJun overexpressing line (Men1-WT-10-JC4) showed highly increased cyclin D1 that remained high on serum starvation.
Compared with Men1-WT-10 vector controls, Men1-NULL-17 and Men1-NULL-41 vector controls showed considerably higher levels of cyclin D1. Unlike Men1-WT-10 overexpressing mJunD, Men1-NULL-17 overexpressing mJunD retained the ability to increase cyclin D1 further on serum stimulation; similarly, in Men1-NULL-41, overexpression of mJunD did not decrease the sustained high levels of cyclin D1 observed under any condition.
Natural JunD Mutations in Tumors. The assembled findings about proliferation, morphology, and cyclin D1 indicate that a JunD missense mutant with disabled JunD-binding to menin is a growth promoter. Endocrine tumors were tested to determine whether similar natural JunD missense mutation could contribute to tumorigenesis. Thirty non-MEN1 parathyroid tumors (20), eleven insulinomas, and six gastrinomas were analyzed for mutations in the N-terminal menin-binding region of JunD (amino acids 1–100). No mutation or polymorphism was detected.
Discussion
The Jun proteins are highly conserved, particularly in their C-terminal basic DNA-binding domain and adjacent bZIP dimerization domain. They are also conserved in several transactivation domains (1). They thus share extensive homology, except in the 95-aa N-terminal region (24). This region has been speculated to account for striking functional differences between JunD and other Jun proteins (25–27). This region, and specifically amino acids 41–44 (in mJunD), is essential for JunD binding to menin (11, 28). The current findings indicate that the contrasting effects of JunD and cJun on growth indices (cell proliferation, cell morphology, and cyclin D1 levels) likely result from their contrasting interactions with menin at or near to these N-terminal sequences.
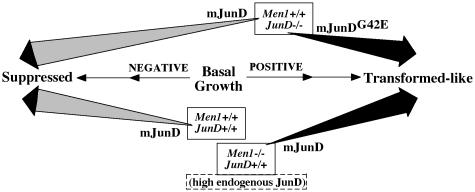
Selected biochemical aspects of menin's repression of JunD transcriptional activity have been explored previously (5, 29, 30). The current studies are directed at JunD's far downstream effects as modified by menin. The effects on any of the three growth indices induced by JunD–/–, Men1–/–, or the overexpression of wt or mutant mJunD in various genotypes are summarized (Fig. 4). Homozygous knockout of either JunD or Men1 alone caused increased growth, supporting prior evidence that either gene might be a growth suppressor (8, 12, 22). The effect of JunD or menin to down-regulate growth indices is not proof for participation of menin in JunD action. However, the fact that this decrease was actually reversed by a missense mutant of JunD unable to bind menin suggests that the growth suppressor effect of JunD depends on a critical function (perhaps binding to menin) involving the mutated JunD codon. Last, the increase in growth indices of menin-null immortalized fibroblasts overexpressing wt mJunD points strongly to menin as the critical partner at or near mJunD codon 42, a partner required for JunD to act as a growth suppressor.
Fig. 4.
Consequence on growth from genotype difference and from stable overexpression of wt or missense mutant mJunD (mJunDG42E is unable to bind menin). The JunD–/– and Men1–/– genotypes are variably displaced to the right from the basal point to reflect their germ-line-positive effect on growth, compared with the wt genotype. Because the JunD-null and the Men1-null immortalized fibroblasts have very different genetic backgrounds, all comparisons between these backgrounds must be qualitative.
The effects of JunD and menin on cyclin D1 expression also paralleled their effects on cell proliferation and morphology. The AP1 site in the cyclin D1 promoter can bind several AP1 proteins, including JunD (31), and could even be the critical site for the direct action of JunD with menin. However, in preliminary experiments, we did not observe any significant effect of menin on mJunD-mediated or mJunDG42E-mediated transactivation of a reporter construct containing a 1.8-kb region of the human cyclin D1 promoter (data not shown). The current experiments do not address whether cyclin D1 is a mediator or a bystander in the JunD-menin actions on growth. Similarly, even an indirect effect of JunD on the cyclin D1 promoter does not affect considerations herein about JunD-menin downstream expressions.
In the current experiments, a transforming-like effect of JunD was unmasked whenever its binding to menin was prevented; i.e., in a menin-null background or in a JunD missense mutant with disabled binding to menin. Menin thus appears to switch JunD's effect on growth toward the opposite direction (Fig. 4). Such a regulatory mechanism as found for JunD could extend, through different processes, to other AP1 proteins. This switching concept arises, in part, from standard terminology for cell growth. This result need not imply a new and opposite activity. More simply put, it could reflect a semantic constraint in defining the true basal point, which could alternatively be conceptualized at the extreme of “suppressed.”
JunD may not normally exist in a “native” form, but only as a complex with menin. To date, menin-null immortalized fibroblasts and certain tumors that are menin-null or menin-inactivated are the only cells known to express JunD without a complex involving menin. Natural mutations or posttranslational changes within the N terminus of JunD could also disrupt the binding to menin. No such mutations have been reported, and none were found on limited testing in the current analysis.
JunD germ-line or somatic mutation has not been proven to contribute to tumorigenesis in vivo. In particular, JunD mutation has not been identified in a human tumor (above). Furthermore, counter to the paradigm of a simple 1:1 pairing of menin and JunD, the phenotypes of Men1 or JunD knockout are very different. That is to say, the Men1–/– mice develop features remarkably similar to those in human MEN1 (18); however, JunD–/– or JunD–/– mice do not develop any MEN1-like, or other tumors (17).
In conclusion, the finding that JunD is converted from a growth suppressor to a growth promoter when it is unable to bind menin provides insights about both JunD and menin. JunD can now be grouped with other AP1 proteins as a positive regulator of cell proliferation. These findings resolve a puzzle that a tumor suppressor (menin) might inactivate another growth suppressor (JunD). It is now evident that JunD and menin together form a growth suppressor unit. This unit can also be formulated as a combination of a growth promoter (native JunD) and a growth suppressor (menin). Established mechanisms for loss of function of this unit include either of two categories of inactivating mutation of menin (i.e., menin loss or disabling of menin's ability to bind to JunD) or missense mutation of JunD that disables JunD binding to menin.
Supplementary Material
Acknowledgments
We thank Dr. Anders Gobl for providing pcDNA3.1-hJunD; Dr. Steven K. Libutti and Dr. Robert T. Jensen for tumors; Dr. Julie Knapp for the mutant JunD plasmids; Dr. George Poy for DNA sequencing; and Dr. Karen Sukhodolets, Dr. Alison Burgess-Hickman, and Dr. Patricia Kennedy for critical reading of the manuscript. In addition, we appreciate the valuable discussions from all of the investigators of the MEN1 collaboration at intramural National Institutes of Health. J.B.W. and M.Y. were supported by grants from the Association for International Cancer Research and the Ligue Nationale Française Contre le Cancer comité de Paris.
Abbreviations: mJunD, mouse JunD; HET, heterozygote; MEN1, multiple endocrine neoplasia type 1; En, embryonic day n; wt, wild type.
References
- 1.Angel, P. E. & Herrlich, P. A. (1994) in The Fos and Jun Family of Transcription Factors, eds. Angel, P. E. & Herrlich, P. A. (CRC, Boca Raton, FL), pp. 3–14.
- 2.Jochum, W., Passegue, E. & Wagner, E. F. (2001) Oncogene 20, 2401–2412. [DOI] [PubMed] [Google Scholar]
- 3.Shaulian, E. & Karin, M. (2001) Oncogene 20, 2390–2400. [DOI] [PubMed] [Google Scholar]
- 4.Agarwal, S. K., Guru, S. C., Heppner, C., Erdos, M. R., Collins, R. M., Park, S. Y., Saggar, S., Chandrasekharappa, S. C., Collins, F. S., Spiegel, A. M., et al. (1999) Cell 96, 143–152. [DOI] [PubMed] [Google Scholar]
- 5.Gobl, A. E., Berg, M., Lopez-Egido, J. R., Oberg, K., Skogseid, B. & Westin, G. (1999) Biochim. Biophys. Acta 1447, 51–56. [DOI] [PubMed] [Google Scholar]
- 6.Chandrasekharappa, S. C., Guru, S. C., Manickam, P., Olufemi, S. E., Collins, F. S., Emmert-Buck, M. R., Debelenko, L. V., Zhuang, Z., Lubensky, I. A., Liotta, L., et al. (1997) Science 276, 404–407. [DOI] [PubMed] [Google Scholar]
- 7.Marx, S. J. (2002) in The Genetic Basis of Human Cancer, eds. Vogelstein, B. & Kinzler, K. W. (McGraw–Hill, New York), pp. 475–450.
- 8.Kim, Y. S., Burns, A. L., Goldsmith, P. K., Heppner, C., Park, S. Y., Chandrasekharappa, S. C., Collins, F. S., Spiegel, A. M. & Marx, S. J. (1999) Oncogene 18, 5936–5942. [DOI] [PubMed] [Google Scholar]
- 9.Poisson, A., Zablewska, B. & Gaudray, P. (2003) Cancer Lett. (Shannon, Irel.) 189, 1–10. [DOI] [PubMed] [Google Scholar]
- 10.Lin, S. Y. & Elledge, S. J. (2003) Cell 113, 881–889. [DOI] [PubMed] [Google Scholar]
- 11.Knapp, J. I., Heppner, C., Hickman, A. B., Burns, A. L., Chandrasekharappa, S. C., Collins, F. S., Marx, S. J., Spiegel, A. M. & Agarwal, S. K. (2000) Oncogene 41, 4706–4712. [DOI] [PubMed] [Google Scholar]
- 12.Pfarr, C. M., Mechta, F., Spyrou, G., Lallemand, D., Carillo, S. & Yaniv, M. (1994) Cell 76, 747–760. [DOI] [PubMed] [Google Scholar]
- 13.Castellazzi, M., Spyrou, G., La Vista, N., Dangy, J. P., Piu, F., Yaniv, M. & Brun, G. (1991) Proc. Natl. Acad. Sci. USA 88, 8890–8894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schreiber, M., Kolbus, A., Piu, F., Szabowski, A., Mohle-Steinlein, U., Tian, J., Karin, M., Angel, P. & Wagner, E. F. (1999) Genes Dev. 13, 607–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weitzman, J. B., Fiette, L., Matsuo, K. & Yaniv, M. (2000) Mol. Cell 6, 1109–1119. [DOI] [PubMed] [Google Scholar]
- 16.Passegue, E., Jochum, W., Behrens, A., Ricci, R. & Wagner, E. F. (2002) Nat. Genet. 30, 158–166. [DOI] [PubMed] [Google Scholar]
- 17.Thepot, D., Weitzman, J. B., Barra, J., Segretain, D., Stinnakre, M. G., Babinet, C. & Yaniv, M. (2000) Development (Cambridge, U.K.) 127, 143–153. [DOI] [PubMed] [Google Scholar]
- 18.Crabtree, J. S., Scacheri, P. C., Ward, J. M., Garrett-Beal, L., Emmert-Buck, M. R., Edgemon, K. A., Lorang, D., Libutti, S. K., Chandrasekharappa, S. C., Marx, S. J., et al. (2001) Proc. Natl. Acad. Sci. USA 98, 1118–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guru, S. C., Goldsmith, P. K., Burns, A. L., Marx, S. J., Spiegel, A. M., Collins, F. S. & Chandrasekharappa, S. C. (1998) Proc. Natl. Acad. Sci. USA 95, 1630–1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heppner, C., Kester, M. B., Agarwal, S. K., Debelenko, L. V., Emmert-Buck, M. R., Guru, S. C., Manickam, P., Olufemi, S. E., Skarulis, M. C., Doppman, J. L., et al. (1997) Nat. Genet. 16, 375–378. [DOI] [PubMed] [Google Scholar]
- 21.Risser, R. & Pollack, R. (1974) Virology 59, 477–489. [DOI] [PubMed] [Google Scholar]
- 22.Okazaki, S., Ito, T., Ui, M., Watanabe, T., Yoshimatsu, K. & Iba, H. (1998) Biochem. Biophys. Res. Commun. 250, 347–353. [DOI] [PubMed] [Google Scholar]
- 23.Bakiri, L., Lallemand, D., Bossy-Wetzel, E. & Yaniv, M. (2000) EMBO. J. 19, 2056–2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Berger, I. & Shaul, Y. (1991) Oncogene 6, 561–566. [PubMed] [Google Scholar]
- 25.Bannister, A. J., Oehler, T., Wilhelm, D., Angel, P. & Kouzarides, T. (1995) Oncogene 11, 2509–2514. [PubMed] [Google Scholar]
- 26.Franklin, C. C., McCulloch, A. V. & Kraft, A. S. (1995) Biochem. J. 305, 967–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Claret, F. X., Hibi, M., Dhut, S., Toda, T. & Karin, M. (1996) Nature 383, 453–457. [DOI] [PubMed] [Google Scholar]
- 28.Yazgan, O. & Pfarr, C. M. (2001) Cancer Res. 61, 916–920. [PubMed] [Google Scholar]
- 29.Gallo, A., Cuozzo, C., Esposito, I., Maggiolini, M., Bonofiglio, D., Vivacqua, A., Garramone, M., Weiss, C., Bohmann, D. & Musti, A. M. (2002) Oncogene 21, 6434–6445. [DOI] [PubMed] [Google Scholar]
- 30.Yumita, W., Ikeo, Y., Yamauchi, K., Sakurai, A. & Hashizume, K. (2003) Int. J. Cancer 103, 738–744. [DOI] [PubMed] [Google Scholar]
- 31.Albanese, C., Johnson, J., Watanabe, G., Eklund, N., Vu, D., Arnold, A. & Pestell, R. G. (1995) J. Biol. Chem. 270, 23589–23597. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.