Abstract
Group I introns are common in the 23 rRNA genes of mitochondria and chloroplasts. Often, they encode “homing endonucleases,” which target highly conserved gene sequences and drive interorganellar intron mobility, even across species and genus lines. Most bacterial 23S rRNA genes show these same endonuclease-sensitive target sequences. However, only two bacterial 23S rRNA genes are known to contain group I introns: that of Simkania negevensis [Everett, K. D., Kahane, S., Bush, R. M. & Friedman, M. G. (1999) J. Bacteriol. 181, 4734–4740], where the intron is not spliced and probably limits growth, and that of Coxiella burnetii [Seshadri, R., et al. (2003) Proc. Natl. Acad. Sci. USA 100, 5455–5460], where no direct evidence of splicing exists. Both bacteria are intracellular parasites and might have acquired introns from eukaryotic hosts. Here we provide direct evidence for splicing, and evolutionary evidence for mobility, of group I introns in the 23S rRNA genes of several free-living hyperthermophilic bacteria of the genus Thermotoga. These bacteria do not live closely with eukaryotes, but phylogenetic analyses suggest that their introns were also acquired from eukaryotic (probably algal) organelles. In vivo, their introns must be spliced at temperatures approaching 90°C, making them the most thermostable natural ribozymes so far described. We demonstrate that at least some of these introns can also self-splice in vitro.
Group I introns constitute a distinct class of ribozyme, characterized by conserved primary and secondary structure and capable of protein-assisted or self-splicing, in vivo and in vitro (1, 2). Many group I introns also carry ORFs encoding “homing endonucleases,” which render them mobile. Homing endonucleases in the 23S rRNA genes of eukaryotic chloroplasts and mitochondria specifically cleave conserved sequences in intron-free 23S rRNA genes. Repair of cleaved genes, templated by intron-containing intact genes, leads to unidirectional “gene conversion,” or copying of the intron, its endonuclease ORF, and flanking sequences into intron-free recipient genes (3, 4). Group I introns can thus move between widely divergent species that retain the endonuclease target sequence, an evolutionary process that has been extensively documented among plant and algal plastids and mitochondria (3, 5, 6). Indeed, it can be convincingly argued that periodic homing provides the only selective pressure for retention of endonuclease activity (7). If this argument is true, then evidence for selection acting on endonuclease gene sequence (for instance, evidence that synonymous codon changes predominate, especially at sites required for activity) can by itself be taken as evidence for evolutionarily recent homing activity.
Until very recently, 23S (or 16S) rDNA introns were unknown in bacteria, a surprise given that most bacterial 23S rRNA genes contain conserved target sequences for intron-encoded homing endonucleases such as I-CeuI and I-CreI (8, 9). Nikolchaeva and Woodson (10) showed that functional or retained (i.e., spliced or unspliced) introns artificially introduced into Escherichia coli 23S rRNA inhibited ribosome formation or function, and Edgell et al. (11) suggested that such deleterious consequences might be one of several “barriers to intron promiscuity in bacteria.”
Group I introns have now been reported in two bacterial 23S rRNA genes, and at least one of them may indeed be deleterious to growth. This intron, described in 1999, is in the 23S rRNA gene of Simikania negevensis (12). It is not spliced out, but persists in the 23S rRNA, where its presence is thought to retard growth (12). The second bacterial 23S rRNA group I intron was only very recently discovered, through the complete sequencing of the genome of Coxiella burnetii (13). Structures of this intron and its ORF encoding a LAGLIDADG homing endonuclease (8, 9) are consistent with splicing and nuclease activity, although neither has been demonstrated. S. negevensis and C. burnetii are both obligate intracellular pathogens, encouraging speculation that they acquired introns from the organelles of eukaryotes.
Methods
DNA from different Thermotoga strains was extracted from frozen cell mass donated by K. O. Stetter (University of Regensburg, Regensburg, Germany) by using the protocol of Charbonnier and Forterre (14). RNA from Thermotoga neapolitana NS-E was extracted from the same cell mass by using the RNeasy Mini Kit (Qiagen, Valencia, CA). Other DNAs were gifts from Y. Takahata (Marine Biotechnology Institute, Kamaishi Laboratories, Kamaishi, Japan), S. L'Haridon and C. Jeanthon (University de Bretagne Occidentale, Brest, France), and M. Madsen and T. Lien (University of Bergen, Bergen, Norway).
Amplification of the intron was carried out by using the following primers: Thermotoga23Sintron.2U, GTGACAAGGCCCTGGCGACT, and Thermotoga23Sintron.275L, GGCATCTTCACCCAGACTGA. Amplifications were carried out in 50 μl final volume containing 20–200 ng of template DNA, 1× PCR buffer, 2.5 mM MgCl2, 0.2 mM dNTP, 1 mM each primer, 0.5–1 unit of Taq DNA polymerase or HiFi Taq DNA polymerase (Invitrogen). The reactions were submitted to an initial denaturation at 93°C for 3 min, and then 30 cycles at 93°C for 30 s, 55–57°C for 30 s, and 72°C for 1.5–2 min. The resulting PCR products were either (i) cloned by using the TOPO TA Cloning Kit (Invitrogen) with an average of five individual clones sequenced with T7 and m13rev, or (ii) cleaned by using Microcon PCR columns (Millipore) and sequenced directly by using the Thermotoga23Sintron.2U and Thermotoga23Sintron.275L primers.
RT-PCR was carried out by using the C. therm. Polymerase for Reverse Transcription in Two-Step RT-PCR Kit (Roche Applied Science) with 60°C synthesis or the Omniscript reverse transcription kit (Qiagen) with 37°C synthesis, and the Thermotoga23Sintron.275L primer in the first step. The second PCR step was carried out as described above with 57°C annealing. The resulting PCR products were cloned by using the TOPO TA Cloning Kit (Invitrogen), and individual clones were sequenced by using T7 and m13rev.
RNA for in vitro splicing was transcribed by T7 RNA polymerase (Fermentas) off PCR product obtained by using vector primers flanking the exon–intron inserts described above (cloned into PCR-2.1 TOPO), with 3 mM NTP and 5 mM MgCl2 to minimize self-splicing during transcription (15). Splicing was initiated by adding 0.2 mM GTP, 25 mM MgCl2, and 1 M KCl (16) to the transcripts, and stopped by chilling on ice. The splicing products were analyzed by RT-PCR as described above.
Transcripts for the time-course analysis of the self-splicing reaction were radioactively labeled by adding 1 μlof[α-32P]GTP (10 μCi/μl; 1 Ci = 37 GBq) in a total reaction volume of 50 μl. In these transcription reactions we used 3 mM CTP, UTP, and ATP and 0.8 mM unlabeled GTP. Splicing was carried out as described above in a total volume of 5 μl, electrophoresis was on a 6% polyacrylamide/6 M urea gel, and visualization was by autoradiography.
Samples for Southern blot hybridization were prepared for electrophoresis by digesting 1–2 μg of genomic DNA with restriction enzymes in a total of 20 μl, and the DNA fragments were separated in a 1% agarose gel. The strains in Fig. 4b and Thermotoga subterranea SL1 were included on the blots. Two samples of each strain were electrophoresed, one cut with EcoRI and one cut with HindIII. The gels were blotted onto positively charged nylon membranes (Roche Diagnostics). A probe made from the PCR product covering both the T. neapolitana NS-E intron and the 23S rDNA was used in the hybridizations. Prehybridization and hybridization were carried out in DIG Easy Hyb (Roche Diagnostics) at 42°C in a rotary oven. Washes were performed in a 0.5× SSC-equivalent buffer at 60°C. The digoxigenin (DIG)-labeled probe was detected by using CDP* (Roche Diagnostics), and exposures were 20 min to 1 h depending on the signal strength.
Fig. 4.
Minimum evolution tree of 16S rRNA sequences and intron sequences. (a) Based on logdet distances estimated from 16S rRNA from all Thermotogales strains included in the study, rooted by Aquifex aeolicus.(b) Based on Kimura 2P distances estimated from 16S rRNA from the strains most closely related to T. neapolitana NS-E. (c) Based on Kimura 2P distances estimated from the intron sequences from E. coli position 1931. All trees were estimated in paup* (19) by using 10 random stepwise additions and TBR branch swapping. Numbers on nodes indicate number of times the node was recovered in 100 bootstrap replicates. Presence of a group I intron in the 23S rRNA is indicated in a and b.
The intron secondary structures were inferred by locating conserved sequences (P4 for Tna.bL1931 and P7-P3-P8 for Tsu.bL1926) in nucleotide blast searches against GenBank, and drawn by using xrna. Phylogenetic analyses were carried out by using phylip (17), tree-puzzle (18), and paup* (19).
Results and Discussion
The 23S group I introns we describe here are in strains of species of the free-living hyperthermophilic bacterial genus Thermotoga. They were discovered in suppressive subtractive hybridization experiments (ref. 20 and unpublished data) designed to identify genes that are present in some strains/species of Thermotoga, but not that of the sequenced Thermotoga maritima (strain MSB8), which has an intron-free 23S rRNA gene. With T. neapolitana NS-E as tester and T. maritima MSB8 as driver (unpublished data), we obtained a 403-bp clone of which base pairs 25–117 showed 60% protein identity (expected by chance 2.0 × 10–12, denoted exp. 2e-12) to a putative group I intron site-specific DNA endonuclease from the chloroplast of the green alga Pterosperma cristatum (GenBank accession no. AAL34315), whereas base pairs 295–403 showed 100% DNA identity to base pairs 2045–2162 of the 23S rRNA of T. maritima MSB8. The junction corresponds to a highly conserved site, strongly suggesting that we had detected a group I intron. In fact, the insertion site (corresponding to position 1931 in the E. coli 23S rRNA) is identical to the insertion site of the intron from P. cristatum (21). Southern blot analyses confirmed that only one 23S rRNA gene exists in T. neapolitana NS-E [and the other Thermotoga strains tested and T. maritima MSB8 (22)], ruling out the possibility that this could be a nonfunctional 23S rRNA gene.
PCR amplification with primers flanking the insertion site showed that the T. neapolitana NS-E 23S intron (denoted Tna.bL1931 according to ref. 23; Fig. 1a) is 699 nt. Computer folding of Tna.bL1931 revealed that it is indeed a group I intron, of subgroup IB (24, 25) (Fig. 1a), as is the P. cristatum intron (21). The intron contains one 489-nt-long ORF (from nucleotide 206 to nucleotide 695) that shows 59% protein identity (exp. 8e-41) to a putative single-LAGLIDADG (26) group I homing endonuclease from the chloroplast of Chlorosarcina brevispinosa (GenBank accession no. AAL34389.1), and high similarity to endonucleases from other green algal group I introns. The endonuclease ORF is inserted into the L8 loop (Fig. 1a), as in other group IB introns, notably the IB4 introns in this position in the 23S rRNA genes in the chloroplasts and mitochondria of green algae, the mitochondria of Acanthamoeba castellanii, and the unspliced intron of S. negevensis (24, 26). The length of the T. neapolitana ORF (163 aa) is similar to that of other known LAGLIDADG endonucleases, and its shows very high similarity to the 152-aa I-CpaI along its entire length (40% identities and 65% similarities). I-CpaI has been shown biochemically be a functional endonuclease, cutting at position 1931 (27). All 149 significant hits in translated blast searches were to putative intron-encoded homing endonucleases from eukaryotes, with the exception of the putative endonuclease from the unspliced group I intron in S. negevensis (GenBank accession no. AAD38228, exp. 2e-29), the ORF in the putative group I intron in C. burnetii (GenBank accession no. AE016960, exp. 2e-17), and an endonuclease (related to intron-encoded endonucleases) from Methanopyrus kandleri that appears to be a free-standing ORF (GenBank accession no. NP_613840, exp. 5e-06). The eight best hits were all from introns found in chloroplasts and mitochondria at the same insertion site as the Thermotoga intron (E. coli 23S position 1931).
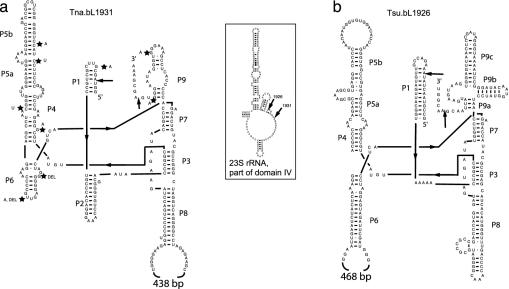
Fig. 1.
Folding of the two Thermotoga introns according to Cech et al. (37). (a) The intron from T. neapolitana NS-E, Tna.bL1931. (b) The intron from T. subterranea SL1, Tsu.bL1926. Sites where polymorphisms were observed among the additional strains that contained an intron similar to Tna.bL1931 are indicated by stars on the structure along with the mutation observed. (Inset) Insertion sites of the two introns relative to part of domain IV of the T. maritima MSB8 23S rRNA, numbered according to the E. coli 23S rRNA. The intron positions in domain IV are found in sequences that form the interface with the 30S ribosome subunit (5).
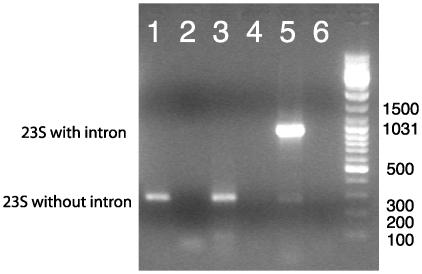
To investigate whether Tna.bL1931 indeed is an intron that is spliced from 23S rRNA, we isolated RNA from T. neapolitana NS-E and performed RT-PCR with primers flanking the insertion site. If the intron is processed out but the exons are not rejoined, no band amplification should be observed, whereas if the intron persists in the 23S rRNA, as observed in S. negevenisis (12), only a 972-bp band should be amplified. However, if the intron is properly spliced (exons rejoined), a 273-bp band is expected. This was indeed what we observed (Fig. 2). The sequence of the 273-bp fragment confirmed that Tna.bL1931 was the spliced intronless rRNA.
Fig. 2.
RT-PCR on RNA isolated from T. neapolitana NS-E. Lane 1, RNA treated with DNase before reverse transcriptase step; lane 2, RNA treated with RNase before reverse transcriptase step; lane 3, RNA; lane 4, no RNA in reverse transcriptase step; lane 5, T. neapolitana NS-E DNA; lane 6, negative control PCR step. A 972-bp fragment in addition to the 273-bp fragment could be amplified from large amounts of fresh RNA (data not shown), suggesting that this PCR product represents either small amounts of DNA contamination or low levels of unspliced rRNA. DNA-dependent PCR on the RNA (i.e., no reverse transcription step) gave only the 972-bp band (data not shown).
This intron must be stable and spliceable in vivo at temperatures at ≈80–90°C. [80°C is optimal and 90°C is maximal temperature for growth of T. neapolitana NS-E (28).] In accordance with this, the G+C content of Tna.bL1931 is much higher than observed for other IB4 introns in the same 23S rRNA location (21): 58% G+C for the intron excluding the ORF and 49% for the complete intron, compared with a G+C content of 29–39% for other intron sequences. High G+C content was also reported for a thermostable group I intron from Azoarcus sp. BH72 tRNAIle (optimum splicing between 70 and 75°C) (29) and a group II intron in the hsp60 gene of Azotobacter vinelandii (optimum splicing at 65°C) (30).
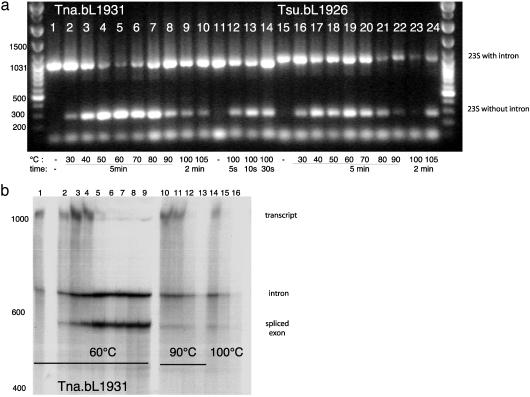
To test whether the intron could self-splice in vitro, we transcribed RNA off the cloned exon–intron fragments, and incubated with Mg2+ and GTP at different temperatures (15). The resulting splicing products were subjected to RT-PCR. If splicing has occurred, both a 972-bp band (intron and exon) and a 273-bp band (exon only) should be amplified. After incubation at various temperatures (30, 40, 50, 70, 80, 90, 100, and 105°C), bands corresponding to the joined exon were indeed detected (Fig. 3a, lanes 1–14). Sequences (data not shown) indicated that some “correct” splicing products were obtained at all temperatures tested, but that some alternatively spliced products also appeared at the higher temperatures. To confirm that in vitro splicing was in fact occurring at elevated temperatures (and not as reaction mixtures were being heated or chilled), the kinetics of product formation with radioactively labeled transcript were monitored at 60, 90, and 100°C (Fig. 3b, lanes 1–16). Reactions were too quick and/or products too unstable at the higher temperatures to permit precise kinetic monitoring, but product formation showed clear time dependence at 60°C.
Fig. 3.
(a) RT-PCR on in vitro splice products obtained by incubating transcripts of the Tna.bL1931 intron (lanes 1–14) and Tsu.bL1926 intron (lanes 15–24) with 273 bp of flanking exon sequence at various temperatures. Lanes 1 and 15, incubated with splicing buffer on ice; lanes 2 and 16, 30°C; lanes 3 and 17, 40°C; lanes 4 and 18, 50°C; lanes 5 and 19, 60°C; lanes 6 and 20, 70°C; lanes 7 and 21, 80°C; lanes 8 and 22, 90°C; lanes 9 and 23, 100°C; lanes 10 and 24, 105°C. Incubation time was 5 min for the incubations at 30–90°C and 2 min for the incubations at 100°C and 105°C. Lane 11, heating the Tna.bL1931 intron–exon transcript to 100°C followed by cooling on ice before addition of splice buffer; lanes 12–14, the Tna.bL1931 intron–exon transcript incubated at 100°C for 5 s (lane 12), 10 s (lane 13), and 30 s (lane 14). (b) Time-course analysis of the self-splicing reaction of Tna.bL1931. RNA was analyzed directly after incubation at 60°C, 90°C, and 100°C at different times. Lane 1, 0 min (incubated with splicing buffer and 0.05 M EDTA on ice); lanes 2–9, incubated at 60°C for 0.5 min (lane 2), 1 min (lane 3), 2 min (lane 4), 5 min (lane 5), 10 min (lane 6), 20 min (lane 7), 30 min (lane 8), 60 min (lane 9); lanes 10–13, incubated at 90°C for 0.5 min (lane 10), 1 min (lane 11), 2 min (lane 12), 5 min (lane 13); lanes 14–16, incubated at 100°C for 0.5 min (lane 14), 1 min (lane 15), and 2 min (lane 16). The full-length transcript is 1,238 nt, the religated exon is 539 nt, and the intron is 699 nt.
To detect introns in other members of the Thermotogales, we screened 18 additional strains (representing five genera and seven species) by using the same flanking primers. Introns were found in nine additional strains (Fig. 4), all in the genus Thermotoga. Six are very close relatives of T. neapolitana NS-E and three have been considered different species. For eight of the introns, very little sequence variation was observed compared with the T. neapolitana NS-E sequence (0.1–11% DNA divergence including the ORF and 0–3% excluding the ORF). All conformed to the same structure as the T. neapolitana NS-E intron and were inserted at the same position in the 23S, and the ORF was inserted in the same intron loop. Phylogenetic analyses showed that the phylogeny resembled that of the 16S rRNA (Fig. 4b), when both excluding and including the ORF (Fig. 4c). The only difference was the position of Thermotoga sp. strain RQ7 and Thermotoga sp. strain SG1. However, the branching pattern in the 16S rRNA tree is not strongly supported, and the intron phylogeny mirrors that obtained from two protein-coding genes (31). Hence, this intron was most likely present in the common ancestor of the Thermotoga strains that possess it presently; it was probably lost from T. maritima MSB8 and Thermotoga sp. strain RQ2 (Fig. 4).
Surprisingly, the intron found in T. subterranea SL1 (denoted Tsu.bL1926; Fig. 1b) proved to be quite different from the T. neapolitana intron, and we infer that it results from an independent acquisition. It is inserted a few base pairs upstream of Tna.bL1931, at position 1926 relative to the E. coli sequence, a position that has been found to harbor nuclear group I introns in different protists (21). Tsu.bL1926 is 774 nt, and computer folding of the intron sequence showed that the structure, although also most similar to group IB introns (25), is different from Tna.bL1931. Tsu.bl1926 lacks P2, has a homing endonuclease ORF inserted into L6, and contains additional P9 helices (Fig. 1b). As observed for Tna.bL1931, the G+C content of this intron is higher than usual (48% excluding the ORF and 42% including the ORF). The G+C content of this intron is nevertheless lower than that of Tna.bL1931, perhaps reflecting that the optimal growth temperature of T. subterranea is 10°C lower than the optimal temperature for T. neapolitana (32), or that it was acquired more recently. The T. subterranea SL1 endonuclease is 507 bp long (inserted between nucleotides 191 and 697) and showed highest similarity to a putative site-specific DNA endonuclease from a group I intron in the mitochondrion of the charophyte Chaetosphaeridium globosum (GenBank accession no. AAM96632, 36% protein identity, exp. 6e-23). The T. subterranea SL1 intron can also be spliced in vitro (Fig. 3a, lanes 15–24). The optimal temperature under the in vitro splicing conditions we used appears to be lower than what we observed for Tna.bL1931 (Fig. 3a). Together with the lower G+C content, these observations are consistent with Tsu.bL1926 being adapted for function at a lower temperature than Tna.bL1931.
A maximum-likelihood tree of the two Thermotoga homing endonucleases together with their closest single-LAGLIDADG matches in blast searches is shown in Fig. 5. Introns and their endonuclease inserted at the same 23S rRNA position have been shown in earlier studies to be more closely related than introns in different positions, even within the same genome (26, 33). The tree in Fig. 5 confirms the close relationship between the endonucleases from all introns at position 1931 in the 23S rRNA and suggests that Tna.bL1931 was acquired from a eukaryote, possibly from the chloroplast or mitochondrion of a green alga. The universally conserved proline residue in α3 (P93 in figure 6 in ref. 26) is replaced by a glutamate residue in all the Tna.bL1931-type ORFs, which might be an adaptation to hyperthermophily (34). The putative endonuclease from T. subterranea is only distantly related to that from Tna.bL1931. The otherwise universally conserved glutamine residue in β2 (Q47 in figure 6 in ref. 26) has been replaced by asparagine in the T. subterranea SL1 endonuclease and in the algal sequences that appear to be most closely related to it: the sequence from Chaetosphaeridium globosum and one of the endonucleases from the chloroplast of Chlorosarcina brevispinosa (c2.Chlorosarcina brevispinosa in Fig. 4, GenBank accession no. AAL34388). None of the other introns reported so far at position 1926 encode LAGLIDADG endonucleases (21), which probably is the reason no very close relative of the Tsu.bL1926 ORF exists. Considering the structural differences (Fig. 1), and the differences in insertion sites between the two introns, it seems certain that group I introns have been acquired at least twice by the Thermotoga lineage.
Fig. 5.
Maximum-likelihood tree of single-LAGLIDADG homing endonucleases carried by group I introns. The 23 first hits to single-LAGLIDADG homing endonucleases in blast-x searches, with the Thermotoga sequences as probes, was selected to build the phylogeny. Chloroplast and mitochondrial introns are indicated by a “c” or an “m,” respectively. Double-LAGLIDADG homing endonucleases were excluded because they have a higher rate of divergence (26). The tree was estimated by using a JTT+Γ model in phylip version 3.6 (17) with a substitution matrix provided by E. Tillier (personal communication), with 10 random additions of the sequences and global rearrangements. The α-parameter was estimated in puzzle version 4.0 (18). Values at nodes indicate number of times the node was recovered in 100 bootstrap replicates (bold numbers) or puzzle support (italic numbers). Insertion position in the 23S rRNA of the group I intron carrying the endonuclease is indicated where this was given in the GenBank entry or available at www.rna.icmb.utexas.edu (21). Specific insertion positions were not available for m. Chlorella vulgaris, m2. Chlorella vulgaris, m. Acanthamoeba castellanii, m2. Acanthamoeba castellanii, and m. Chaetosphaeridium globosum.
Notably, blast searches with the 170 nt of 23S rRNA sequence upstream of the insertion site of T. neapolitana NS-E give two cyanobacteria and a chloroplast 23S rRNA as the first hits after T. maritima MSB8 and Fervidobacterium islandicum, and about half of the 50 best hits were to cyanobacteria or chloroplasts. This pattern was not observed when searching with the whole sequenced fragment or with the 100 nt downstream of the insertion site. This pattern could be due to flanking region co-conversion at the time of intron acquisition (4), and if so, implies that the Fervidobacterium lineage also once harbored such introns.
The data presented here raise additional intriguing questions about the transfer and maintenance of these introns and their encoded endonucleases. Lateral transfer (cross-species homing) of group I introns is well documented among eukaryotes (4, 5) and seems by far the most plausible scenario for initial acquisition of the Thermotoga introns described here. Transfers across species lines and much larger phylogenetic distances have been observed before, but in most cases a shared niche or some physically intimate relationship between the donor and the recipient can be inferred (3, 12). All members of the Thermotoga lineage characterized are free-living, and all are hyperthermophilic or at least thermophilic (optimum growth, 66–80°C) (35). Presumably, they acquired their introns by means of free DNA, viruses, or plasmids encountered where they grow. The very high similarity of the TnaL.b1931 to endonucleases from eukaryotic introns at the same position, together with the fact that the largest reservoirs of group I introns are the nuclei and organelles of eukaryotes (21), suggests that these introns were acquired from a eukaryote. Although it is easy to imagine that hyperthermophiles could acquire DNA from mesophiles unfortunate enough to fall into their special environments, it seems unlikely that a mesophilic endonuclease would function, or a that mesophilic intron would be properly spliced, in a hyperthermophilic background. Thermophilic (but not hyperthermophilic) eukaryotes are known (36), and, of course, an intron and endonuclease functioning marginally at the lower limits of a thermophile's range might subsequently adapt to higher temperatures.
Of the 100 substitutions observed among the nine Tna.b1931 endonucleases we have sequenced, at most 19 would introduce an altered amino acid (average ds/dn in pairwise comparisons = 23.02), suggesting that all are (or have until recently been) under selection for activity at the temperatures at which the organism now encoding them grow. Only two of the nonsynonymous substitutions would affect conserved positions in the internal segment, which covers all but one of the secondary elements in I-CreI (26). Both of these are conservative replacements (I ↔ V and K ↔ R), found in other endonucleases that cut the same site. Because strong theoretical arguments exist that endonuclease activity is selectively maintained only by and through homing (7), we infer that introns have been actively moving in genus Thermotoga since their introduction, before the diversification of the species studied here. More active within-species than between-species transfer is likely, however, because striking incongruence does not occur between phylogenies for endonuclease and 16S rRNA.
If physiological “barriers to intron promiscuity” do exist that explain the paucity of group I introns in other bacterial 23S rRNA genes, they have been relaxed within species of this genus of free-living hyperthermophiles. Thus these introns, in addition to providing models for high-temperature ribozymology and examples for interdomain transfer, might provide a window into hyperthermophile cell and population biology.
Acknowledgments
We are grateful to Dr. Murray Schnare for discussions and help with drawing the intron structures in XRNA, Dr. Amanda Lohan for critical reading of the manuscript, and Marlena Dlutek for excellent technical assistance. We also thank Dr. Karl O. Stetter, Dr. Yoh Takahata, Dr. Stéphane L'Haridon, Dr. Christian Jeanthon, Dr. Marit Madsen, and Dr. Torleiv Lien for donating Thermotogales DNA or cell mass. This work was supported by a postdoctoral fellowship (to C.L.N.) from the Canadian Institute for Health Research and by funds from the Canadian Institutes for Health Research (MOP 4467) and Genome Canada (Genome Atlantic).
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AJ556784–AJ556793).
References
- 1.Cech, T. R. (1990) Annu. Rev. Biochem. 59, 543–568. [DOI] [PubMed] [Google Scholar]
- 2.Cech, T. R. (2001) Biochem. Soc. Trans. 30, 1162–1166. [DOI] [PubMed] [Google Scholar]
- 3.Cho, Y., Qiu, Y. L., Kuhlman, P. & Palmer, J. D. (1998) Proc. Natl. Acad. Sci. USA 95, 14244–91424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lambowitz, A. M. & Belfort, M. (1993) Annu. Rev. Biochem. 62, 587–622. [DOI] [PubMed] [Google Scholar]
- 5.Jackson, S., Cannone, J., Lee, J., Gutell, R. & Woodson, S. (2002) J. Mol. Biol. 323, 35–52. [DOI] [PubMed] [Google Scholar]
- 6.Turmel, M., Otis, C. & Lemieux, C. (2002) Mol. Biol. Evol. 19, 24–38. [DOI] [PubMed] [Google Scholar]
- 7.Goddard, M. R. & Burt, A. (1999) Proc. Natl. Acad. Sci. USA 96, 13880–13885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marshall, P. & Lemieux, C. (1992) Nucleic Acids Res. 20, 6401–6407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jurica, M. S., Monnat, R. J., Jr., & Stoddard, B. L. (1998) Mol. Cell 2, 469–476. [DOI] [PubMed] [Google Scholar]
- 10.Nikolcheva, T. & Woodson, S. A. (1997) RNA 3, 1016–1027. [PMC free article] [PubMed] [Google Scholar]
- 11.Edgell, D. R., Belfort, M. & Shub, D. A. (2000) J. Bacteriol. 182, 5281–5289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Everett, K. D., Kahane, S., Bush, R. M. & Friedman, M. G. (1999) J. Bacteriol. 181, 4734–4740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Seshadri, R., Paulsen, I. T., Eisen, J. A., Read, T. D., Nelson, K. E., Nelson, W. C., Ward, N. L., Tettelin, H., Davidsen, T. M., Beanan, M. J., et al. (2003) Proc. Natl. Acad. Sci. USA 100, 5455–5460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Charbonnier, F. & Forterre, P. (1995) in Archaea: A Laboratory Manual, eds. Robb, F. T. & Place, A. R. (Cold Spring Harbor Lab. Press, Plainview, NY), Vol. 3, pp. 87–90. [Google Scholar]
- 15.Shub, D. A., Peebles, C. L. & Hampel, A. (1994) in RNA Processing: A Practical Approach, eds. Higgins, S. J. & Hames, B. D. (Oxford Univ. Press, Oxford), Vol. 2, pp. 211–239. [Google Scholar]
- 16.Johansen, S. & Vogt, V. M. (1994) Cell 76, 725–734. [DOI] [PubMed] [Google Scholar]
- 17.Felsenstein, J. (2001) phylip Phylogeny Inference Package (Dept. of Genetics, Univ. of Washington, Seattle), Version 3.6.
- 18.Strimmer, K. & von Haeseler, A. (1996) Mol. Biol. Evol. 13, 964–969. [Google Scholar]
- 19.Swofford, D. L. (2001) paup*, Phylogenetic Analysis Using Parsimony (*and Other Methods) (Sinauer, Sunderland, MA), Version 4.
- 20.Nesbø, C. L., Nelson, K. E. & Doolittle, W. F. (2002) J. Bacteriol. 184, 4475–4488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cannone, J. J., Subramanian, S., Schnare, M. N., Collett, J. R., D'Souza, L. M., Du, Y., Feng, B., Lin, N., Madabusi, L. V., Muller, K. M., et al. (2002) BMC Bioinformatics 3, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nelson, K. E., Clayton, R. A., Gill, S. R., Gwinn, M. L., Dodson, R. J., Haft, D. H., Hickey, E. K., Peterson, J. D., Nelson, W. C., Ketchum, K. A., et al. (1999) Nature 399, 323–329. [DOI] [PubMed] [Google Scholar]
- 23.Johansen, S. & Haugen, P. (2001) RNA 7, 935–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Michel, F. & Westhof, E. (1990) J. Mol. Biol. 216, 585–610. [DOI] [PubMed] [Google Scholar]
- 25.Cech, T. R. (1988) Gene 73, 259–271. [DOI] [PubMed] [Google Scholar]
- 26.Lucas, P., Otis, C., Mercier, J. P., Turmel, M. & Lemieux, C. (2001) Nucleic Acids Res. 29, 960–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Turmel, M., Cote, V., Otis, C., Mercier, J. P., Gray, M. W., Lonergan, K. M. & Lemieux, C. (1995) Mol. Biol. Evol. 12, 533–545. [DOI] [PubMed] [Google Scholar]
- 28.Jannasch, H. W., Huber, R., Belkin, S. & Stetter, K. O. (1988) Arch. Microbiol. 150, 103–104. [Google Scholar]
- 29.Tanner, M. & Cech, T. (1996) RNA 2, 74–83. [PMC free article] [PubMed] [Google Scholar]
- 30.Adamidi, C., Fedorova, O. & Pyle, A. M. (2003) Biochemistry 42, 3409–3418. [DOI] [PubMed] [Google Scholar]
- 31.Nesbø, C. L., L'Haridon, S., Stetter, K. O. & Doolittle, W. F. (2001) Mol. Biol. Evol. 18, 362–375. [DOI] [PubMed] [Google Scholar]
- 32.Jeanthon, C., Reysenbach, A. L., L'Haridon, S., Gambacorta, A., Pace, N. R., Glenat, P. & Prieur, D. (1995) Arch. Microbiol. 164, 91–97. [PubMed] [Google Scholar]
- 33.Turmel, M., Otis, C., Cote, V. & Lemieux, C. (1997) Nucleic Acids Res. 25, 2610–2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cambillau, C. & Claverie, J. M. (2000) J. Biol. Chem. 275, 32383–32386. [DOI] [PubMed] [Google Scholar]
- 35.Reysenbach, A.-L. (2001) in Bergeys Manual of Systematic Bacteriology, eds. Boone, D. R., Castenholz, R. W. & Garrity, G. M. (Springer, New York), 2nd Ed., Vol. 1, pp. 369–385. [Google Scholar]
- 36.Baumgartner, M., Stetter, K. O. & Foissner, W. (2002) J. Eukaryotic Microbiol. 49, 227–238. [DOI] [PubMed] [Google Scholar]
- 37.Cech, T. R., Damberger, S. H. & Gutell, R. R. (1994) Nat. Struct. Biol 1, 273–280. [DOI] [PubMed] [Google Scholar]