Abstract
Although horizontal gene transfer is well documented in microbial genomes, no case has been reported in higher plants. We discovered horizontal transfer of the mitochondrial nad1 intron 2 and adjacent exons b and c from an asterid to Gnetum (Gnetales, gymnosperms). Gnetum has two copies of intron 2, a group II intron, that differ in their exons, nucleotide composition, domain lengths, and structural characteristics. One of the copies, limited to an Asian clade of Gnetum, is almost identical to the homologous locus in angiosperms, and partial sequences of its exons b and c show characteristic substitutions unique to angiosperms. Analyses of 70 seed plant nad1 exons b and c and intron 2 sequences, including representatives of all angiosperm clades, support that this copy originated from a euasterid and was horizontally transferred to Gnetum. Molecular clock dating, using calibrations provided by gnetalean macrofossils, suggests an age of 5 to 2 million years for the Asian clade that received the horizontal transfer.
Horizontal gene transfer is the basis for the genetic engineering of commercially important crops, and natural horizontal gene transfers across kingdoms have been documented between Agrobacterium and Nicotiana (1), Wolbachia and its insect host Callosobruchus (2), and bacteria and land plants (3, 4). Among eukaryotic lineages, however, very few natural horizontal transfers have been reported, and none of them involve transfers across groups of seed plants. As part of an investigation of the phylogeny of Gnetum (Gnetales, gymnosperms), we studied the distribution of a group II intron in the mitochondrial (mt) nad1 gene, which encodes subunit 1 of the respiratory chain NADH dehydrogenase. The nad1 gene consists of five exons that are cisor trans-located and separated by 7–294 kb (refs. 5–8 and Fig. 1A). The intervening introns are cis- or trans-spliced accordingly (9, 10). The second intron of the nad1 gene, located between exons b and c, is a group II intron (ref. 11 and Fig. 1 A). Group II introns are self-splicing RNAs that are typical components of contemporary organellar genomes in plants, algae, fungi, protists, and eubacteria (10, 12–14). They are characterized by a uniform structure of six major domains radiating from a central wheel (Fig. 1B), with domains I and V most crucial for the introns' enzymatic activity (12, 14) and ribozymic function (11, 12). Because of these characteristics and the presence of reverse transcriptase ORFs, they are believed to be the ancestors of spliceosomal introns and non-long-terminal repeat retroelements (13, 15). Unlike group I introns, at least one of which appears to have been traded within flowering plants (16), group II introns in plants have been thought to be strictly vertically inherited (17–19), and the only known horizontal transfer of a group II intron in eukaryotes occurred in haptophytes, marine unicellular flagellates (20).
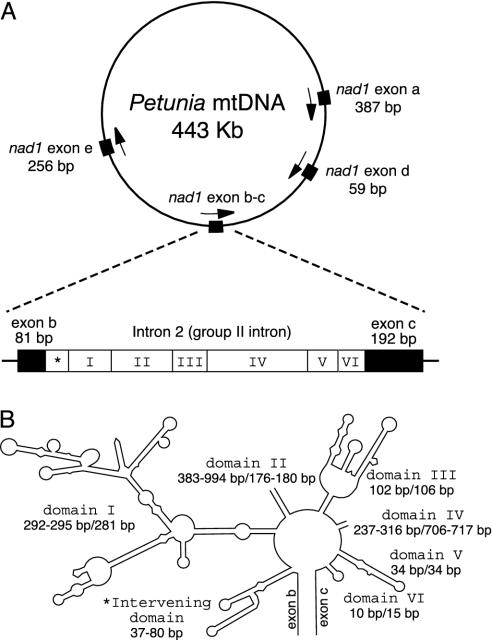
Fig. 1.
Relative location of the five nad1 exons in the Petunia mt genome and a predicted secondary structure of the gymnosperm-type mt nad1 intron 2 of Gnetum gnemonoides.(A) Arrows indicate the 5′ → 3′ orientation of the nad1 exon regions. An asterisk in the intron 2 scheme indicates the intervening nucleotides of the Gnetum gymnosperm-type intron. Mitochondrial map modified from Conklin et al. (6). (B) Domain lengths before the slash refer to Gnetum gymnosperm-type introns, those after the slash to Gnetum angiosperm-type introns.
During work on the phylogeny of Gnetum, we developed specific primers to amplify the second intron in the nad1 gene, plus flanking exons, from numerous previously unstudied species of Gnetum. We discovered that Gnetum harbors two copies of nad1 intron 2 and reasoned that a larger-scale survey of complete nad1 intron 2 sequences should allow identification of the source of the different copies because major seed plant lineages have specific intron signatures. If the origin of the two copies in Gnetum could be traced, this might provide evidence for horizontal gene transfer, adding a new dimension to our understanding of group II intron evolution. An earlier survey of the distribution of nad1 intron 2 in 276 angiosperms and 17 gymnosperms representing all major lineages of seed plants had shown that the intron is widespread, but absent in seven unrelated angiosperms, non-Pinaceae conifers, and the Gnetales genus Welwitschia (19). The sister group of Welwitschia, Gnetum, was found to have the intron, whereas the third genus of Gnetales, Ephedra, gave a weak hybridization signal and multiple PCR products. Given its apparent multiple loss in angiosperms, the intron's distribution in gymnosperms was most parsimoniously explained by parallel independent losses in Welwitschia and the ancestor of non-Pinaceae conifers (19). This interpretation of a few repeated losses, rather than multiple regains, is independent of the specific relationships among seed plants and the mono- or paraphyly of Coniferales, which are currently unresolved (ref. 21 and references therein). However, these previous studies of the distribution of nad1 intron 2 in seed plants have mainly relied on Southern hybridization.
Materials and Methods
Taxon Sampling. We obtained 38 accessions of Gnetum, other Gnetales, and angiosperms; these are listed together with voucher information and GenBank accession numbers in Table 2. Most samples were collected in the field by H.W., but a few are from botanical gardens or herbarium material. Partial exon b and c sequences are available from a range of seed plants, and full intron sequences for 18 additional angiosperm taxa were generated in our lab (Table 2).
Gene Sequencing. We extracted DNA from silica gel-dried leaves by using Qiagen (Valencia, CA) Plant DNeasy mini kits. Concentration and quality of extracted DNAs were checked by 1.5% agarose gel electrophoresis with a Lambda/HindIII/EcoRI size marker. We amplified the entire nad1 intron 2 region by using the Expand High Fidelity PCR system (Roche Applied Science), using primers “exonB” and “exonC” designed by Demesure et al. (ref. 22; Figs. 4 and 5, which are published as supporting information on the PNAS web site). The 5′ and 3′ region sequences of each type were compared with partial Gnetum gnemon sequences (19), and their homologues were searched via blastn searches. Exon sequences homologous to partial G. gnemon sequences were designated as “gymnosperm-type,” whereas exon sequences homologous to angiosperm ones were designated “angiosperm-type.” Type-specific internal primers were then designed to prevent amplification of the other sequence type (Figs. 4 and 5), and appropriate amounts (0.5–4 ng) of template DNA were added to the PCR master mixture following the manufacturer's protocol. Portions of the PCR products were run on an 1.5% agarose gel with a Lambda/HindIII/EcoRI size marker to verify the size and amount of the PCR products (Fig. 5). Single band PCR products were purified with Qiagen PCR purification kits. Double bands were separated via 1.2% agarose gel electrophoresis and then purified using Qiagen QIAEX II gel purification kits. Separated and purified PCR products were PCR-sequenced, using ABI Prism Big Dye Terminator Cycle Sequencing Ready Reaction kits (Perkin–Elmer) with the amplification primers. Sequences were aligned and cleaned-up in seqman ii (DNASTAR).
Sequence Analysis and Secondary Structure Comparison. Gnetum mt nad1 intron 2 and partial exon b and c sequences were compared with homologous regions in Citrullus (watermelon, Cucurbitaceae) whose secondary structure has been predicted (11). This helped to find and align the stem regions of the six domains. A data matrix of seed plant nad1 exon b and c and intron 2 sequences was constructed that included information on the domain regions, by using se-al software (version 2.0a11; Fig. 2 and Fig. 6, which is published as supporting information on the PNAS web site). We verified the secondary structure by the thermodynamic method using mfold software (version 3.1, www.bioinfo.rpi.edu/applications/mfold/; refs. 23 and 24; Fig. 1B). Domains between ID and I3 are excluded from Fig. 6 because of mismatches in the thermodynamically predicted secondary structure with the Citrullus model (Fig. 1B). Length and GC content of each domain were calculated from the aligned sequences (Tables 3 and 4, which are published as supporting information on the PNAS web site). Sequence divergences were calculated by using the General Time Reversible model with among-site rate variation and proportion of invariant sites (GTR + G + I; ref. 25) estimated by Bayesian analysis (Table 1 and Table 5, which is published as supporting information on the PNAS web site).
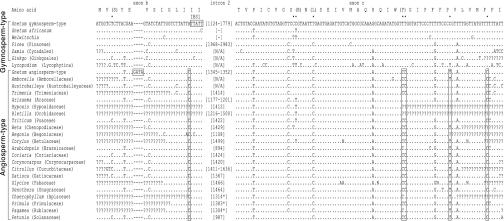
Fig. 2.
Alignment of mt nad1 exons b and c. Dots indicate identity to topmost sequence; dashes indicate gaps; question marks indicate missing data. Numbers in brackets indicate lengths of intron; N/A and an asterisk after the intron length indicate incomplete intron sequences; dashes indicate lack of intron. Amino acid translation is given for Gnetum gymnosperm-type exon sequences, and amino acids that have undergone RNA editing (5–7) are given in parentheses. Dots above the topmost sequence indicate the posttranscriptionally edited sites. The specific Gnetum gymnosperm-type and angiosperm-type introns included are those of G. klossii. The Gnetum angiosperm-type nad1 exon b is characterized by a 4-nt (GATA) insertion. IBS1 indicates the intron binding site. See Table 2 for GenBank accession numbers.
Table 1. Mitochondrial nad1 intron 2 sequence divergences based on the GTR + G + I model with α = 1.036 and Pinv = 0.079.
| Trimenia | Monocots | Rosids | Asterids | |
|---|---|---|---|---|
| Gnetum angiosperm-type | 0.12-0.15 | 0.11-0.18 | 0.06-0.15 | 0.03-0.09 (0.03-0.06)* |
| Trimenia (basal angiosperm) | 0.10-0.19 | 0.10-0.19 | 0.08-0.15 | |
| Monocots | 0.11-0.24 | 0.09-0.18 | ||
| Rosids | 0.06-0.14 |
Sequence divergence between Gnetum and Pagamea and Petunia (Asterids).
Phylogenetic Analysis. Phylogenetic analyses used paup* version 4.0b10 (26) and mr. bayes (27). For Fig. 3A, partial sequences of mt nad1 exons b and c (Fig. 2) and the conserved sections of intron 2 (Fig. 6) from 85 accessions (Table 2) were analyzed by neighbor-joining, with sequence divergences calculated under the GTR + G + I model (25) after removing all gaps and ambiguous characters. Nonparametric bootstrap support was obtained by resampling the data 1,000 times with the same search options and model. Parsimony analyses of full intron sequences using heuristic searching were run separately for each intron type. Heuristic searching used 100 random taxon addition replicates, holding 100 trees at each step, tree bisection–reconnection branch swapping, MulTrees, Collapse, and Steepest Descent options, and no upper limit for trees held in memory. The partial exons/intron sequence tree and two full-length intron sequence trees had basically identical topologies, with the intron trees showing more phylogenetic structure.
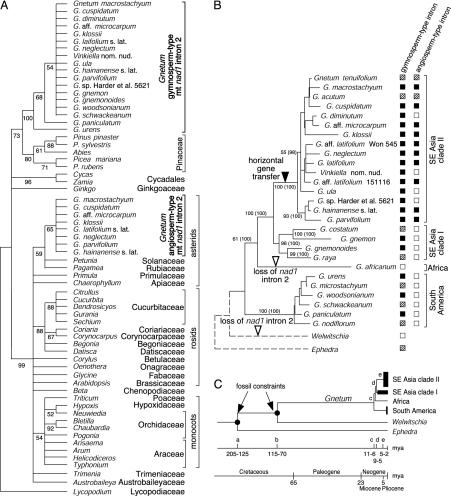
Fig. 3.
Distribution of mt nad1 partial exons b and c and intron 2, and age estimates for Gnetum crown groups. (A) Bootstrap majority-rule consensus tree obtained from conserved intron and exon sections analyzed by neighbor-joining, using GTR + G + I distances (α = 0.097 and Pinv = 0.120). (B) Distribution of the gymnosperm- and angiosperm-type introns on the maximum likelihood tree obtained from concatenated Gnetum nuclear and plastid sequences. Filled squares indicate intron presence; hatched squares represent partially sequenced or PCR-verified accessions; open squares indicate lack of intron. No introns were detected in G. africanum and Welwitschia. Numbers next to nodes represent parsimony bootstrap support, followed by Bayesian posterior probabilities. (C) Age estimates for Gnetum crown groups from chloroplast matK and rbcL sequences. Arrows indicate nodes constrained by fossils. a, age of Gnetales (205–125 my, depending on the fossil used); b, age of the Gnetum-Welwitschia divergence (115–70 my); c, age of extant Gnetum (11–6 my); d, age of Asian Gnetum; e, age of Asian clade II Gnetum.
To construct a phylogenetic framework in which to analyze the distribution of the nad1 intron 2 within Gnetum, we sequenced five additional loci (together 4,540 aligned base pairs): the nuclear Leafy gene second intron, the nuclear internal transcribed spacers/5.8S, the plastid rbcL gene, the plastid matK gene, and the plastid tRNALeu (UAA) intron and adjacent spacers. Combined data were analyzed, after removing all gaps and ambiguous sites, under parsimony, maximum likelihood, and Bayesian optimization. The final data set included 40 accessions of Gnetum, of which 27 nonredundant ones are included in Fig. 3B to represent the species recognized in a new monograph (H.W., unpublished data). Parsimony analysis used the same options as above. Maximum likelihood used the GTR + Gamma + Pinv model. Bayesian probabilities were obtained under the GTR + Gamma model, with four Markov chain Monte Carlo chains run for 556,700 generations, using random trees as starting points, sampling every 10th generation, and discarding the first 16,300 trees as burn-in.
Age Estimation of Crown Gnetum Lineages. For the age estimates, combined chloroplast matK and rbcL sequences of seed plants were analyzed under maximum likelihood, using the GTR + Gamma + Pinv model. Psilotum was included to root seed plants. Likelihood ratio tests were used to assess clock-like behavior of the data. Branch lengths in Gnetales were calibrated with Gurvanella (28), a fossil consisting of branches and attached seeds, and Cratonia cotyledon, a seedling macrofossil from the Brazilian Crato formation (29, 30). Gurvanella constrains the minimum age of Gnetales to 125 million years (my) ago (Barremian–Aptian) because it combines the characteristic mode of branching of Ephedra with the seed morphology of Welwitschia; the seedling from the Crato formation shows an embryo feeder, epidermis and venation characters unique to Gnetum and Welwitschia, and constrains the minimum age of their most recent common ancestor to 115 my ago (Aptian–Albian).
Results
Sequence Characteristics of Gymnosperm- and Angiosperm-Type Exons/Intron. Development of primers specific to Gnetum gymnosperm-type and angiosperm-type nad1 intron 2 sequences was possible because they possess unique characteristics. The gymnosperm-type nad1 intron 2 in Gnetum is characterized by 37–80 base pairs inserted before the GTGCG motif that is typical of the start of group II introns (Tables 3 and 4 and Figs. 1 and 6). Angiosperm-type nad1 exons found in Gnetum share five nucleotide substitutions with angiosperms that are not found in other seed plants. They are also characterized by a four-nucleotide (GATA) insertion that makes exon b nonfunctional (Fig. 2). The uniqueness of the two types of Gnetum mt nad1 intron 2 sequences precludes contamination as an explanation for our findings. As shown by Gugerli et al. (19) for Gnetum gnemon, the 37–80 base pairs are not translated. By using primers specific for each intron type and sequencing entire introns together with parts of exons b and c, we confirmed (i) the presence of gymnosperm-type introns and exons in all accessions of Gnetum except Gnetum africanum and (ii) the additional presence of angiosperm-type introns and exons in 19 accessions of Gnetum representing at least 10 species (Table 2 and Figs. 3 and 6).
The length of the gymnosperm-type introns in Gnetum ranged from 1,124 to 1,779 bp, whereas that of the angiosperm-type introns ranged from 1,345 to 1,352 bp (Tables 3 and 4). Length variation in Gnetum gymnosperm-type introns was due mainly to domain II, which comprised between 383 and 994 bp. Domain IV varied between 237 and 316 bp, and domains I (292–295 bp), III (102 bp), V (34 bp), and VI (10 bp) were invariant in length across accessions (Fig. 1B). As expected from this length variation, gymnosperm-type introns across seed plants were difficult to align. Thus, in domain IV, Pinaceae introns were up to 10 times longer than Gnetum introns (1,347–2,367 vs. 237–316 bp), whereas in domain II, Gnetum introns were up to six times longer than Pinaceae introns (383–994 vs. 159 bp). Angiosperm-type introns showed little variation in domain lengths except for domain IV (Tables 3 and 4) and were thus relatively easily alignable. As expected from these differences, alignment of the Gnetum gymnosperm-type intron sequences with Gnetum angiosperm-type intron sequences was not feasible except in the stem regions (Figs. 1B and 6).
Sequence divergences between the nad1 intron of angiosperms and the angiosperm-type intron of Gnetum (Tables 1 and 5) lie between 0.03 and 0.18, with the closest similarity (0.03–0.06) to Pagamea (Rubiaceae) and Petunia (Solanaceae), members of the euasterid I clade (31–33).
Phylogenetic Position of Gymnosperm- and Angiosperm-Type Exons/Intron. Fig. 3A shows the tree resulting from the analysis of partial exons and conserved sections of the nad1 intron 2 sequences obtained from Gnetum and other seed plants. Gnetum gymnosperm-type nad1 exons b and c and intron sequences cluster with Pinaceae, whereas Gnetum angiosperm-type sequences cluster inside the flowering plants. A Gnetum/Pinaceae clade is relatively well supported (73% bootstrap support) as found in other studies (refs. 19 and 21 and references therein), and intra-Gnetum relationships are congruent with those resulting from combined nuclear and chloroplast markers in that the South American species are sister to the Southeast Asian and African species (Fig. 3B). Angiosperm-type exon and intron sequences, including those from Gnetum, group together with 99% bootstrap support (lower part of Fig. 3A), and the Gnetum sequences are closest to the euasterids Pagamea (Rubiaceae) and Petunia (Solanaceae). The relationships among the remaining angiosperm nad1 introns are generally congruent with current three- and five-gene phylogenies for the same taxa (31–33).
Plotting of the two types of exons/introns on a phylogeny of Gnetum (Fig. 3B) reveals that the angiosperm-type sequences are restricted to one of two Southeast Asian clades (labeled “clade II”). Gnetum species from South America and the remainder of Southeast Asia contain only gymnosperm-type exons/introns, and the single African species, G. africanum, lacks either intron type. Strikingly, the angiosperm-type exons/introns are lacking, and apparently were lost secondarily, in four species of clade II (Fig. 3B).
Age Estimation of Crown Gnetum Lineages. Because a likelihood ratio test of models with and without a globally optimal substitution rate did not reject the clock assumption (0.05 > P > 0.01), branch lengths in chloroplast matK+rbcL data were calibrated with gnetalean macrofossils (Materials and Methods). Calibration with the Gurvanella fossil yielded a divergence time of 7 to 6 my for all extant Gnetum species and of 5 to 2 my for species in the Asian clade II of Gnetum (Table 6, which is published as supporting information on the PNAS web site, and Fig. 3C). Calibration with the Cratonia cotyledon fossil yielded an age of Gnetales of 205 to 184 my, of crown Gnetum of 11 to 10 my, and of the Asian clade II of 4 my.
Discussion
Origin of the Foreign Exon/Intron Copies Found in Gnetum. The presence of angiosperm-like nad1 intron 2 and adjacent exons in one clade of Gnetum, in addition to the presence of a gymnosperm-like intron 2 and adjacent exons b and c in all gnetums except the African G. africanum, necessitates abandonment of the prevailing view that group II introns are strictly parentally inherited in seed plants and instead suggests horizontal transfer of this piece of genome from a flowering plant to a gymnosperm. The loss of intron 2 (gymnosperm-type?) in G. africanum, the only African species of Gnetum, is striking because the closest relative of Gnetum, Welwitschia, with a sole species, Welwitschia mirabilis, also from Africa, is one of very few other seed plants that lack an nad1 intron 2 (19). This raises the possibility that a gymnosperm-type intron 2 was lost in the common ancestor of Gnetum and Welwitschia, and regained within Gnetum. Although the retrohoming/homing and coconversion of group II introns have been reported for yeast (34, 35) and horizontal transfer of group II introns has been reported from marine unicellular flagellates (20), we know of no case from seed plants in which a group II intron was lost and then regained (see also ref. 19). Unlike the group II introns of prokaryotes and lower eukaryotes, organellar group II introns of land plants apparently have lost ORFs that could encode endonucleases and retrotranscriptases essential for retrohoming (12, 14). The group II intron studied here, mt nad1 intron 2, lost its ORF completely, and only fragmentary sequence homology between Arabidopsis thaliana mt nad1 intron 2 and Marchantia polymorpha mt atp9 intron 1 suggests past presence of an ORF (14).
The phylogenetic distribution of the angiosperm-type intron in Gnetum suggests that the horizontal gene transfer happened in the stem lineage of the Asian clade II (Fig. 3B). Different from the gymnosperm-type introns, which show large divergences in accordance with their long evolutionary history, the angiosperm-type introns show little divergence and size variation (Tables 1 and 3–5), indicative of short divergence times. Based on a local molecular clock, species in Gnetum clade II diverged from each other only 5 to 2 my ago (Fig. 3C). Relatively recent horizontal gene transfer from a Southeast Asian asterid angiosperm to the common ancestor of Gnetum clade II thus is the most parsimonious explanation for the distribution of the two copies of nad1 intron 2 found in Gnetum.
An alternative hypothesis explaining the observed phylogenetic incongruence between the two (xenologous) types of Gnetum nad1 introns would involve persistence of an ancient seed plant nad1 intron 2 polymorphism, followed by selective extinction (36, 37). Such an explanation requires that the nad1 second intron and adjacent exon sections duplicated in the most recent common ancestor of angiosperms and Gnetales and that the angiosperm-type intron and exons b and c then survived only in Gnetum and angiosperms. The age of the most recent common ancestor of angiosperms and Gnetales is currently unknown and depends on the resolution of seed plant phylogenetic relationships. However, the low sequence divergence between nad1 intron 2 in Pagamea and Petunia vs. Gnetum (0.03–0.06) argues against an ancient divergence of these introns. Also, all Gnetum angiosperm-like introns cluster with asterids (Pagamea and Petunia), that is, derived angiosperms, rather than as a sister lineage to angiosperms, but the age of the angiosperm nad1 intron 2 is thought to predate the split of monocots and dicots some 140 my ago (10). The monophyly of angiosperm sequences, including the Gnetum angiosperm-type exons/introns, leaves no other possible explanation but horizontal gene transfer.
Possible Mechanisms of Horizontal Gene Transfer. The mechanisms by which horizontal gene transfer takes place are largely a matter of speculation; agents that have been implied are viruses, bacteria, fungi, and plant cell-piercing insects (see ref. 3). The discovery of mt heteroplasmy in Silene acaulis (38) suggests the possibility of transfer of an entire angiosperm mitochondrion to Gnetum during insect (moth or fly) pollination (39, 40). Mechanisms involved in yeast group II intron retrohoming/homing and coconversion (34, 35) are unlikely to apply to the present case of horizontal transfer because seed plant nad1 second introns have lost the ORFs crucial for retrohoming. Instead, the presence of angiosperm-specific characteristics in the upstream and downstream exons of Gnetum angiosperm-type nad1 intron 2 (Fig. 2), and especially that these sites are posttranscriptionally edited (RNA editing) in angiosperms (refs. 5–7 and Fig. 2), suggest that the exons and intron have transferred simultaneously as DNA. The insertion of the GATA motif in the Gnetum angiosperm-type exon b may have occurred during the horizontal transfer or immediately after the transfer, because all Gnetum angiosperm-type sequences share this insert. Extra indels in angiosperm-type exon b occur in Gnetum aff. latifolium SAN151116 and Gnetum neglectum (data not shown), further supporting the pseudogenization of this exon. We found no evidence for pseudogenization in exon c, nor did we find evidence in the intron for secondary structure disruption.
Future work is needed to clarify the subcellular location of the angiosperm-type exons/introns as well as the scope of the horizontal transfer (see below). Resolving the location will require mapping of the Gnetum mt genome, which would also clarify the arrangement of the five exons of the nad1 gene (Fig. 1 A). So far, intact group II introns have not been found in nuclear genomes (12), and incorporation of mt introns or genes into chloroplasts appears extremely rare (41, 42). That the horizontally transferred group II intron and its flanking exons may still be located in mitochondria is also suggested by the absence of acceleration in the substitution rates of the Gnetum angiosperm-type intron. Genes transferred from mitochondria to nuclei often acquire accelerated substitution rates (18, 41, 43, 44).
The scope of the horizontal transfer clearly involved the intron and adjacent exons. An involvement of all five exons (a–e) of the nad1 gene is unlikely because of their wide separation (Fig. 1 A). The presence of two identical copies of mt nad1 exon a in maize (8) and of a group II intron in mt nad5 in Huperzia selago, which in addition has a group II intron in its mt nad1 (45), demonstrates that several copies of nad exons or introns can coexist in single mt genomes.
The extent and mechanisms of horizontal gene transfer between eukaryotes are not well understood, with concomitant unease about the release of genetically modified organisms. As shown here, horizontal transfer of mt DNA segments has occurred naturally between gymnosperms and angiosperms in their recent evolutionary past, and Bergthorsson et al. (46) reported instances of such transfers within flowering plants. These results indicate that natural mechanisms exist for the horizontal transfer of mt genes, suggesting that horizontal gene transfer may play an underestimated role in the evolution of seed plants.
Supplementary Material
Acknowledgments
H.W. thanks the International Center for Tropical Ecology at the University of Missouri (St. Louis) for financial support of his fieldwork, and herbaria and field collectors for leaf samples. We thank J. D. Palmer for his critical reading and insightful suggestions and anonymous reviewers for their helpful comments.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: mt, mitochondrial; my, million years.
Data deposition: The DNA sequences reported in this paper have been deposited in the GenBank database (accession nos. AY230269–AY230316, AY231296–AY231300, AY243113, AY243121, AY243125, AY243129–AY243131, AY256880–AY256885, and AY283607–AY283610). For a full listing of accession numbers, see Table 2, which is published as supporting information on the PNAS web site, www.pnas.org.
References
- 1.Aoki, S. & Syõno, K. (1999) Proc. Natl. Acad. Sci. USA 96, 13229–13234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kondo, N., Nikoh, N., Ijichi, N., Shimada, M. & Fukatsu, T. (2002) Proc. Natl. Acad. Sci. USA 99, 14280–14285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Syvanen, M. & Kado, C. I., eds. (2002) Horizontal Gene Transfer (Academic, London), 2nd Ed.
- 4.Brown, J. R. (2003) Nat. Rev. Genet. 4, 121–132. [DOI] [PubMed] [Google Scholar]
- 5.Chapdelaine, Y. & Bonen, L. (1991) Cell 65, 465–472. [DOI] [PubMed] [Google Scholar]
- 6.Conklin, P. L., Wilson, R. K. & Hanson, M. R. (1991) Genes Dev. 5, 1407–1415. [DOI] [PubMed] [Google Scholar]
- 7.Wissinger, B., Schuster, W. & Brennicke, A. (1991) Cell 65, 473–482. [DOI] [PubMed] [Google Scholar]
- 8.Fauron, C. M.-R., Moore, B. & Casper, M. (1995) Plant Sci. 112, 11–32. [Google Scholar]
- 9.Malek, O., Brennicke, A. & Knoop, V. (1997) Proc. Natl. Acad. Sci. USA 94, 553–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Malek, O. & Knoop, V. (1998) RNA 4, 1599–1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Michel, F., Umesono, K. & Ozeki, H. (1989) Gene 82, 5–30. [DOI] [PubMed] [Google Scholar]
- 12.Michel, F. & Ferat, J.-L. (1995) Annu. Rev. Biochem. 64, 435–461. [DOI] [PubMed] [Google Scholar]
- 13.Lambowitz, A. M., Caprara, M. G., Zimmerly, S. & Perlman, P. S. (1999) in The RNA World: The World of Modern RNA Suggests a Prebiotic RNA, eds. Gesteland, R. F., Cech, T. R. & Atkins, J. F. (Cold Spring Harbor Lab. Press, Plainview, NY), 2nd Ed., pp. 451–485.
- 14.Toor, N., Hausner, G. & Zimmerly, S. (2001) RNA 7, 1142–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zimmerly, S., Guo, H., Perlman, P. S. & Lambowitz, A. M. (1995) Cell 82, 545–554. [DOI] [PubMed] [Google Scholar]
- 16.Cho, Y., Qiu, Y.-L., Kuhlman, P. & Palmer, J. D. (1998) Proc. Natl. Acad. Sci. USA 95, 14244–14249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qiu, Y.-L., Cho, Y., Cox, J. C. & Palmer, J. D. (1998) Nature 394, 671–674. [DOI] [PubMed] [Google Scholar]
- 18.Palmer, J. D., Adams, K. L., Cho, Y., Parkinson, C. L., Qiu, Y.-L. & Song, K. (2000) Proc. Natl. Acad. Sci. USA 97, 6960–6966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gugerli, F., Sperisen, C., Büchler, U., Brunner, I., Brodbeck, S., Palmer, J. D. & Qiu, Y.-L. (2001) Mol. Phyl. Evol. 21, 167–175. [DOI] [PubMed] [Google Scholar]
- 20.Ehara, M., Watanabe, I. K. & Ohama, T. (2000) Gene 256, 157–167. [DOI] [PubMed] [Google Scholar]
- 21.Magallón, S. & Sanderson, M. J. (2002) Am. J. Bot. 89, 1991–2006. [DOI] [PubMed] [Google Scholar]
- 22.Demesure, B., Sodzi, N. & Petit, R. J. A. (1995) Mol. Ecol. 4, 129–131. [DOI] [PubMed] [Google Scholar]
- 23.Mathews, D. H., Sabina, J., Zuker, M. & Turner, D. H. (1999) J. Mol. Biol. 288, 911–940. [DOI] [PubMed] [Google Scholar]
- 24.Zuker, M., Mathews, D. H. & Turner, D. H. (1999) in RNA Biochemistry and Biotechnology, eds. Barciszewski, J. & Clark, B. F. C. (Kluwer Academic, Dordrecht, The Netherlands), pp. 11–43.
- 25.Swofford, D. L., Olsen, G. J., Waddell, P. J. & Hillis, D. M. (1996) in Molecular Systematics, eds. Hillis, D. M., Moritz, C. & Mable, B. K. (Sinauer, Sunderland, MA), 2nd Ed., pp. 430–459.
- 26.Swofford, D. L. (2002) PAUP*: Phylogenetic Analysis Using Parsimony (*and Other Methods) (Sinauer Associates, Sunderland, MA), version 4.
- 27.Huelsenbeck, J. P. & Ronquist, F. R. (2001) Bioinformatics 17, 754–755. [DOI] [PubMed] [Google Scholar]
- 28.Zhou, Z., Barrett, P. M. & Hilton, J. (2003) Nature 421, 807–814. [DOI] [PubMed] [Google Scholar]
- 29.Mohr, B. A. & Friis, E. M. (2000) Int. J. Plant Sci. 161, Suppl. 6, 155–167. [Google Scholar]
- 30.Rydin, C., Mohr, B. A. & Friis, E. M. (2003) Proc. R. Soc. London Ser. B 270, Suppl., 29–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qiu, Y.-L., Lee, J., Bernasconi-Quadroni, F., Soltis, D. E., Soltis, P. S., Zanis, M., Zimmer, E. A., Chen, Z., Savolainen, V. & Chase, M. W. (1999) Nature 402, 404–407. [DOI] [PubMed] [Google Scholar]
- 32.Soltis, P. S., Soltis, D. E. & Chase, M. W. (1999) Nature 402, 402–404. [DOI] [PubMed] [Google Scholar]
- 33.Soltis, D. E., Soltis, P. S., Chase, M. W., Mort, M. E., Albach, D. C., Zanis, M., Savolainen, V., Hahn, W., Hoot, S. B., Fay, M. F., et al. (2000) Bot. J. Linn. Soc. 133, 381–461. [Google Scholar]
- 34.Eskes, R., Liu, L., Ma, H., Chao, M. Y., Dickson, L., Lambowitz, A. M. & Perlman, P. S. (2000) Mol. Cell. Biol. 20, 8432–8446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Belfort, M., Derbyshire, M., Parker, M., Cousineau, B. & Lambowitz, A. M. (2002) in Mobile DNA II, eds. Craig, N. L., Craigie, R., Gellert, M. & Lambowitz, A. M. (Am. Soc. Microbiol. Press, Washington, DC), pp. 761–783.
- 36.Cummings, M. P. (1994) Trends Ecol. Evol. 9, 141–145. [DOI] [PubMed] [Google Scholar]
- 37.Wendel, J. F. & Doyle, J. J. (1998) in Molecular Systematics of Plants II: DNA Sequencing, eds. Soltis, D. E., Soltis, P. S. & Doyle, J. J. (Kluwer Academic, Boston), pp. 265–296.
- 38.Städler, T. & Delph, L. F. (2002) Proc. Natl. Acad. Sci. USA 99, 11730–11735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kato, M. & Inoue, T. (1994) Nature 368, 195. [Google Scholar]
- 40.Kato, M., Inoue, T. & Nagamitsu, T. (1995) Am. J. Bot. 82, 862–868. [Google Scholar]
- 41.Adams, K. L., Daley, D. O., Qiu, Y.-L., Whelan, J. & Palmer, J. D. (2000) Nature 408, 354–357. [DOI] [PubMed] [Google Scholar]
- 42.Adams, K. L., Qiu, Y.-L., Stoutemyer, M. & Palmer, J. D. (2002) Proc. Natl. Acad. Sci. USA 99, 9905–9912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wolfe, K. H., Li, W. S. & Sharp, P. M. (1987) Proc. Natl. Acad. Sci. USA 84, 9054–9058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Laroche, J., Li, P., Maggia, L. & Bousquet, J. (1997) Proc. Natl. Acad. Sci. USA 94, 5722–5727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vangerow, S., Teerkorn, T. & Knoop, V. (1999) Plant Biol. 1, 235–243. [Google Scholar]
- 46.Bergthorsson, U., Adams, K. L., Thomason, B. & Palmer, J. D. (2003) Nature 424, 197–201. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.