Abstract
Previous work has demonstrated a role for the E2F1 gene product in signaling apoptosis, both as a result of the deregulation of the Rb/E2F pathway as well as in response to DNA damage. We now show that the ability of cells to suppress the apoptotic potential of E2F1, as might occur during the course of normal cellular proliferation, requires the action of the Ras–phosphoinositide 3-kinase–Akt signaling pathway. In addition, we also identify a domain within the E2F1 protein, previously termed the marked-box domain, that is essential for the apoptotic activity of E2F1 and that distinguishes the E2F1 protein from E2F3. We also show that the E2F1-marked-box domain is essential for the induction of both p53 and p73 accumulation. Importantly, a role for the marked-box domain in the specificity of E2F1-mediated apoptosis coincides with recent work demonstrating a role for this domain in achieving specificity in the activation of transcription. We conclude that the unique capacity of E2F1 to trigger apoptosis reflects a specificity of transcriptional activation potential, and that this role for E2F1 is regulated through the action of the Akt protein kinase.
The role of the Rb/E2F pathway in the regulation of cell cycle progression, particularly the G1/S transition, is now well established (1, 2). A variety of experiments have suggested distinct roles for individual members of the E2F family in the control of cell proliferation and cell fate (1, 2). For example, various experiments have suggested a particularly important role for E2F3 in the control of cell proliferation, consistent with a role in the activation of genes encoding DNA replication proteins (3, 4). In contrast, E2F4 may not be so critical for proliferation, but rather it may foster the ability of various cell types to exit the cell cycle and differentiate (5, 6). E2F1 would appear to play dual roles in the control of cell proliferation and cell fate. Overexpression of E2F1 can drive quiescent cells into S phase (7), and studies of cells deleted for multiple E2F proteins provide evidence of a role for E2F1 in the ability of cells to proliferate (8). But, in addition to this role, E2F1 appears to have the unique ability to induce apoptosis when expressed in the absence of proliferative signals (9–14).
A physiological role for the E2F1-induced apoptosis has been seen in several contexts. E2F1–/– mice develop thymic hyper-plasia caused by a defect in thymocyte apoptosis (15), and the absence of E2F1 activity has been associated with decreased negative selection of T cells (16). In addition, mice lacking Rb function exhibit excessive apoptosis in the developing neurons, lens, and myocytes (17, 18). At least part of the Rb–/– phenotype is caused by E2F1 deregulation (19–21), but it is also true that other studies have provided evidence of a role for E2F3 in the Rb-dependent apoptosis pathway (22). Additional studies have shown that E2F1 is uniquely induced in response to DNA damage (23). The specificity of the response reflects the fact that E2F1, but not the other E2Fs, is a direct target for the ATM (ataxia telangiectasia mutated) and ATR (ATM and Rad3-related) protein kinases, which are induced after DNA damage. ATM/ATR-mediated phosphorylation of E2F1 leads to an accumulation of the protein caused by inhibition of the normal degradation of the E2F1 protein.
These observations, pointing to the specificity of function within the E2F family and also the dual role of E2F1 in signaling both apoptosis and cell proliferation, raise the question of the mechanism of this specificity. What distinguishes E2F1 from the other E2F proteins in triggering the apoptotic pathway, and how is this E2F1-induced apoptosis blocked during normal cell proliferation? The induction of the apoptotic pathway is known to be associated with an ability of E2F1 to induce the accumulation of p53, as well as various other genes encoding apoptotic activities. The induction of p53 accumulation has now been shown to involve the activation of p19ARF protein, which then controls Mdm2 function. Mdm2 has been shown to control p53 protein levels by targeting p53 for ubiquitin-mediated degradation (24). The link with E2F1 is now seen from the observation that E2F induces p19ARF expression, likely a direct transcription activation via E2F sites in the ARF promoter, and with an apparent specificity for E2F1 (14, 25). Moreover, the activation of genes such as p73 and Apaf1 has also been suggested to be an E2F1-specific process (26, 27). E2F1 can also block apoptosis by repressing the expression of proapoptotic genes such as Mcl-1 and Traf2 (28, 29).
By using a series of chimeric proteins that interchange E2F1 and E2F3 domains, we demonstrate that the marked-box domain of E2F1 is critical for the ability of E2F1 to specifically induce apoptosis. Moreover, we further show that the suppression of E2F1-induced apoptosis during normal growth depends on the activity of the Ras-dependent Akt pathway.
Materials and Methods
Cells and Viruses. REF52 cells were passaged in medium containing 10% serum (50% fetal calf serum and 50% bovine calf serum). Adenoviruses were constructed by using the AdEasy system designed by He et al. (30). Viral titers were determined by indirect immunofluorescence against the viral Mr 72,000 E2 gene product. Titers for all adenoviruses used in these studies were determined simultaneously to avoid experimental variation. REF52 cells plated on 60-mm dishes were infected with adenovirus at the stated multiplicities of infection (mois) in 500 μl of DMEM containing no serum for 1.25 h at 37°C with frequent rocking of the plates. After infection, medium containing the noted serum concentration was added to each plate.
Caspase-3 Cleavage Assays. To bring REF52 cells to quiescence, cells were plated in serum-containing medium for 24 h before washing and adding back starvation medium containing 0.25% serum. Cells were serum starved for 48 h before infection with adenovirus. For assays performed in the presence of serum, cells were plated in 10% serum-containing medium for 24 h before infection with adenovirus. The cells were plated at an initial density such that at the time of infection, the plates each contained ≈3 × 105 cells per 60-mm diameter dish. Before infection, cells from one serum-deprived and one serum-containing dish were counted, so equivalent mois could be used. Floating and adherent cells were processed for caspase-3 cleavage detection 40 h after infection, following the manufacturer's protocol (catalog no. 550914, BD PharMingen).
Construction of Chimeric E2Fs. The cDNAs of hemagglutinin (HA)-E2F1 and HA-E2F3 were mutagenized to introduce restriction recognition sites to facilitate the cloning of individual domains between these genes. An NruI site was introduced into HA-E2F1 and HA-E2F3 between the N-terminal cyclin A-binding region and the DNA-binding domain. We inserted a ScaI site into E2F1 and E2F3 between the DNA-binding and DP1 dimerization sites. We introduced a BstBI site into E2F1 that aligned with a naturally occurring site in E2F3 and separates the DP1 dimerization and “marked-box” regions. A PvuI site was introduced into HA-E2F3 that aligned with a naturally occurring site in E2F1 to separate the marked-box from the “marked-box-adjacent” region. A PmlI site was introduced into both E2F1 and E2F3, serving to separate the marked-box-adjacent region from the C-terminal transactivation/pocket protein domain.
Immunoblot Analysis. REF52 cells were harvested 24 h after infection into microcentrifuge tubes and resuspended in boiling sample buffer. Equivalent amounts of protein were separated by SDS/PAGE, transferred to an Immobilon-P (Millipore) membrane, and blocked in T-TBS (20 mM Tris/0.137 M NaCl/0.2% Tween 20) containing 5% nonfat dry milk. Blots were then incubated with primary antiserum (1:1,000) at room temperature for 4 h, washed three times with T-TBS, and then incubated with the appropriate secondary antiserum (1:2,000) for 1 h at room temperature. Blots were processed by using the ECL Western Blotting System (Amersham Biosciences). For p73 westerns, Saos-2 cells were deprived of serum for 48 h and infected with the indicated viruses expressing E2F chimeras. Nuclear extracts were prepared 24 h after infection and analyzed by immunoblotting. Antiserum against HA (sc-805) and actin (sc-8432) from Santa Cruz Biotechnology and p53 (OP09) and p73 (OP108) from Oncogene were used for Western blot analysis.
5-Bromodeoxyuridine (BrdUrd) Incorporation Assays. Assays were performed as described (31).
Results
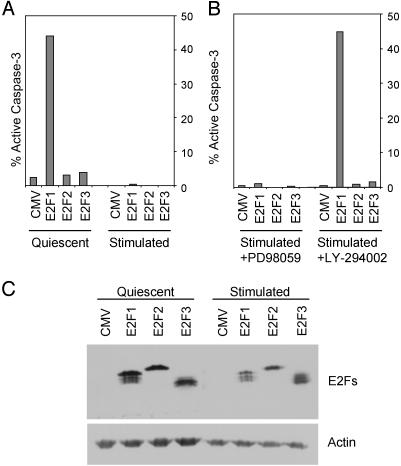
A Role for Phosphoinositide 3-Kinase (PI3K)/Akt in Suppressing E2F1-Induced Apoptosis. Previous work has demonstrated a role for E2F1 in triggering apoptosis when expressed in the absence of survival signals provided by serum (9, 10). This role also coincides with an ability of E2F1 to induce the accumulation of p53 protein (11) as well as the expression of several proapoptotic genes (26, 32). A number of studies have provided evidence that this activity is specific for E2F1 (11, 14), although there are instances where other E2Fs, particularly E2F3, also appear to contribute to apoptosis (22). A further example of the E2F1 specificity in induction of apoptosis can be seen in the assay shown in Fig. 1, where E2F1, but not E2F2 or E2F3, was capable of inducing cell death of quiescent fibroblasts, as measured by caspase-3 cleavage, a direct downstream effector of normal apoptosis (Fig. 1 A). In this experiment, we have used E2F proteins tagged with HA to compare the accumulation of the three proteins. Western blotting against the HA epitope revealed that the E2Fs were expressed at roughly equivalent levels. An actin Western blot demonstrated equal protein loading among the lanes (Fig. 1C).
Fig. 1.
A serum-activated PI3K-signaling pathway blocks E2F1-induced apoptosis. (A) E2F1-induced apoptosis is blocked by serum. REF52 cells were deprived of serum (quiescent) or grown in serum (stimulated) as described in Materials and Methods. REF52 cells were infected on the same day with E2F-expressing adenoviruses at an moi of 75 focus-forming units per cell. CMV, cytomegalovirus control. Samples were harvested 40 h after infection and assayed for active caspase-3 levels by fluorescence-activated cell sorter (FACS; Becton Dickinson) analysis. (B) REF52 cells were grown in 10% serum-containing medium for 24 h. After infection with the indicated E2F adenoviruses, medium containing 10% serum and either 100 μM mitogen-activated protein kinase kinase (MEK) inhibitor (PD 98059) or 50 μM PI3K inhibitor (LY-294002) was added back to the cells. Cells were harvested 40 h after infection and processed for active caspase-3 levels by FACS analysis. (C) Equal expression of E2F-producing HA-E2Fs. Protein extracts were prepared from REF52 cells infected with E2F adenoviruses 24 h after infection. Extracts were analyzed by Western blotting using antiserum against HA (E2Fs) or actin to verify equal protein loading.
The fact that E2F1 accumulation is a normal consequence of the passage of cells through G1/S after growth stimulation suggests that the apoptotic signal must be suppressed during normal cell proliferation. Indeed, the addition of serum to cells expressing E2F1 can be seen to completely suppress the apoptotic activity (Fig. 1 A). Serum stimulation activates a large number of intracellular signaling cascades, in which Ras is centrally involved. Activation of Ras has been implicated in the control of normal cellular growth as well as contributing to the aberrant proliferation of cancer cells. Multiple Ras effector pathways have been identified, including the Raf/Mek/Erk signaling pathway, which has been demonstrated to directly lead to transcriptional activation of the Mdm2 gene (33). This pathway is thought to function as an antiapoptotic signal, because Mdm2 directly targets p53 for proteasomal degradation. Ras can also block apoptosis through the direct activation of the PI3K signaling pathway. Active PI3K indirectly leads to activation of protein kinase B (PKB)/Akt, a serine/threonine kinase that phosphorylates and inactivates a large number of apoptosis-inducing genes. Because active Ras can deliver antiapoptotic signals through both the MEK and PI3K signaling pathways, we explored the role of these pathways in controlling the E2F1-induced apoptosis.
As shown in Fig. 1B, the MEK inhibitor PD 98059 did not alter the capacity of serum to block E2F1-induced apoptosis, although it did block Erk phosphorylation (data not shown). In contrast, addition of the PI3K inhibitor LY-294002 completely abrogated the serum-mediated suppression of E2F1 apoptosis. Importantly, even in the absence of PI3K activity, E2F2 and E2F3 still failed to induce apoptosis, further underscoring the unique role of E2F1 in this function. We thus conclude that E2F1 does exhibit a unique ability to trigger apoptosis, and that this ability is regulated by a PI3K-dependent signaling pathway associated with the stimulation of normal cell growth.
Hyperactive Akt Attenuates E2F1-Induced Apoptosis and the Induction of p53. PI3K catalyzes the phosphorylation of plasma-membrane-localized phosphoinositides that then serve as docking platforms for downstream signaling molecules, including the Akt/PKB (34). After Akt relocalizes to the plasma membrane, it is phosphorylated and activated by the membrane associated PDK1 kinase. Various studies have shown that active Akt can block apoptosis by phosphorylating and inactivating components of the cellular apoptotic machinery, such as Bad, caspase-9, and the forkhead transcription factors. Akt also phosphorylates I-KKα, causing the release of NF-κB, a transcription factor important for cell survival (35). We have thus explored the role of Akt to suppress E2F1-induced apoptosis.
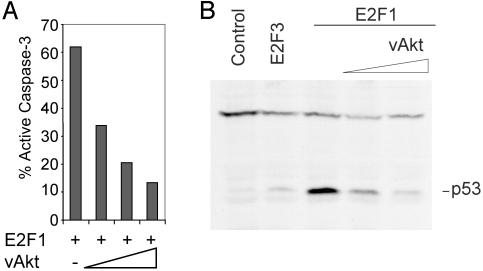
The viral Akt gene recovered in the transforming retrovirus AKT8 represents a fusion of viral gag sequences with the cellular akt gene, resulting in a constitutively active vAkt protein that is targeted to the plasma membrane by the viral gag sequence independent of Ras–PI3K signaling (36, 37). We generated a recombinant adenovirus containing the v-Akt gene and used this virus to coexpress active Akt together with E2F1. Approximately 60% of cells infected with Ad-E2F1 displayed an apoptotic phenotype, as measured by caspase-3 cleavage (Fig. 2A). Coinfection of the cells with the Ad-Akt virus led to a dose-dependent reduction in the number of apoptotic cells, resulting in an 80% decrease in apoptosis at the highest levels of vAkt.
Fig. 2.
Constitutively active Akt attenuates E2F1-induced apoptosis and p53 accumulation. (A) Constitutively active Akt blocks E2F1-induced apoptosis in REF52 cells. Serum-deprived REF52 cells were infected with E2F1 adenovirus-expressing (150 moi) and increasing mois (150, 300, and 500) of vAkt-producing adenovirus. Cells were harvested 40 h after infection and assayed for active caspase-3. (B) Constitutively active Akt blocks E2F1-dependent p53 protein accumulation. Serum-starved REF52 cells were infected at an moi of 150 with control (CMV), E2F3-, E2F1-, or E2F1-expressing adenoviruses, along with increasing amounts of adenovirus that expresses hyperactive Akt (moi 150 and 300). A control adenovirus (vector lacking an insert) was used in place of the Akt virus for the samples shown in lanes 3 (moi 300) and 4 (moi 150). Whole cell lysates were prepared 24 h after infection, and equivalent protein amounts from each infection were separated by SDS/PAGE and probed with anti-p53 antiserum.
Although the majority of known Akt substrates control apoptosis downstream of p53, the finding that vAkt can block E2F1-induced apoptosis prompted us to explore whether this blocking may be, in part, fostered by an ability of vAkt to block E2F1-induced p53 protein accumulation. Serum-starved REF52 cells were infected with control virus (CMV) or Ad-E2F1 along with increasing levels of vAkt. As a control, cells were also infected with the Ad-E2F3 virus. Whole cell extracts were prepared 24 h after infection and p53 levels were measured by immunoblot. As shown in Fig. 2B, E2F1 but not E2F3 induced the accumulation of p53 protein. Importantly, vAkt blocked the E2F1-dependent p53 protein accumulation. This blocking demonstrates that Akt can influence apoptosis by targeting molecules upstream of p53 accumulation. This finding is in accordance with recently published reports stating that Akt can directly bind to, phosphorylate, and activate murine Mdm2 and human Hdm2. This phosphorylation causes Mdm2/Hdm2 to shuttle from the cytoplasm to the nucleus, where it targets p53 for degradation, thus promoting cell survival (38–41)
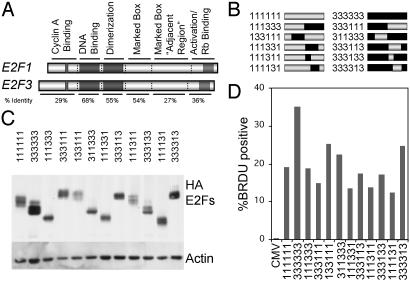
Identification of E2F1 Sequences Responsible for Induction of Apoptosis. The data presented here, together with previous work, provide evidence of a unique role for E2F1 in the induction of apoptosis. We have taken advantage of this differential activity to identify a domain within E2F1 that is required for the induction of apoptosis. To determine the domain(s) that allow E2F1 but not E2F3 to induce apoptosis, a set of chimeric E2F cDNAs was constructed. cDNAs encoding HA-tagged E2F1 and E2F3 were used for these studies to allow immunodetection with a single antibody. After E2F1 and E2F3 sequences were aligned, suitable restriction enzyme sites were introduced into the cDNAs by site-directed mutagenesis. These silent mutagenic sites were chosen to allow the cloning of individual E2F domains between the cDNAs without perturbing the conserved domains.
We focused on six regions within the two E2F proteins, based on previous work that has highlighted functional roles for these domains. The first represents the N terminus, which has been shown to regulate protein accumulation, the second denotes the DNA-binding domain, the third is the DP1 dimerization domain, the fourth represents the so-called marked-box domain, the fifth is a region adjacent to the marked-box domain, and the sixth is the transactivation/Rb-binding domain (Fig. 3A). For reference in the chimeras, a “1” or a “3” denotes the origin of the domain as either E2F1 or E2F3, respectively (Fig. 3B). Each of the chimeras was expressed from an adenovirus vector to facilitate expression of the E2Fs in cells. Serum-starved REF52 cells were infected with chimeric E2Fs at an moi of 75, the same moi used for the apoptotic assays, and returned to serum starvation media for 24 h. Whole cell extracts were prepared and analyzed by SDS/PAGE. A Western blot assay with HA antiserum revealed that the chimeric E2Fs were expressed at roughly equivalent levels (Fig. 3C). To verify that each of the chimeras was functional, we tested their ability to induce quiescent cells to enter S phase. REF52 cells were deprived of serum for 48 h, infected with chimeric Ad-E2Fs, and returned to 0.25% serum medium for 8 h before adding 50 μM BrdUrd for 15 h. Cells were fixed, and BrdUrd labeling was observed by indirect immunofluorescence with anti-BrdUrd antibody (Amersham Biosciences). As seen in Fig. 3D, each of the chimeras induced significant levels of BrdUrd incorporation compared with control virus (CMV), indicating that each of the chimeras is expressed and is functional.
Fig. 3.
Construction of chimeric E2Fs. (A) Restriction sites were introduced into the cDNAs of HA-E2F1 and HA-E2F3 to allow the cloning of individual domains between the genes. The introduced restriction sites are outlined in Materials and Methods. (B) Diagram of the chimeric E2Fs described in this study. The nomenclature describes the identity of individual domains in the chimeras: first digit, N terminus/cyclin A binding; second digit, DNA-binding domain; third digit, DP1 dimerization domain; fourth digit, marked box; fifth digit, marked-box-adjacent region; sixth digit, transactivation/Rb-binding domain. Resultant E2F cDNAs were used to generate recombinant adenovirus expressing the HA-E2Fs. (C) HA-E2F protein expression in REF52 cells. Serum-starved REF52 cells were infected with the noted HA-E2F-expressing adenovirus and processed for Western blotting 24 h after infection. Equal amounts of protein were separated by SDS/PAGE and probed with anti-HA antiserum. Actin is shown as a loading control. (D) Induction of BrdUrd incorporation. Serum-starved REF52 cells were infected with the chimeric E2F-expessing adenoviruses and returned to 0.25% serum-containing medium for 8 h. Cells were then labeled with 50 μM BrdUrd for 15 h, and BrdUrd incorporation was assayed by indirect immunofluorescence.
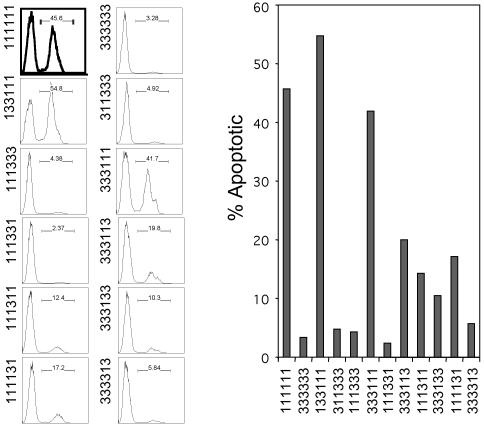
Quiescent REF52 cells were infected with adenovirus expressing chimeric E2Fs at an moi of 75 focus-forming units per cell and returned to medium containing 0.25% serum. Cells were harvested 40 h after infection, and levels of active, cleaved caspase-3 were measured by flow cytometric analysis. Comparable to the results in Fig. 1, ≈45% of the cells expressing E2F1 undergo apoptosis, whereas expression of E2F3 did not induce apoptosis (Fig. 4). Expression of the chimera containing the E2F3 DNA-binding domain and DP1 dimerization domain (133111) was able to induce apoptosis as strongly as E2F1, demonstrating that the specificity for E2F1-mediated apoptosis does not reside within the DNA-binding domain. This conclusion is validated by the observation that the reverse chimera (311333) failed to induce apoptosis, and by the fact that the chimera containing only the C-terminal half of E2F1 (333111) also induced apoptosis as strongly as E2F1, whereas the reverse chimera (111333) was inactive. These results point to the C-terminal sequences as mediating the apoptotic function and exclude both the N-terminal region and the DNA-binding domain.
Fig. 4.
The E2F1 marked-box domain is required for the induction of apoptosis. (Left) REF52 cells were incubated in 0.25% serum medium for 48 h and then infected with the indicated chimeric E2F-producing adenovirus at an moi of 75 focus-forming units per cell. After infection, cells were returned to starvation medium for 40 h before harvesting for detection of active caspase-3 levels by fluorescence-activated cell sorter (FACS; Becton Dickinson) analysis. (Right) The flow cytometry data shown in Left are presented here in graph format.
To further define the region in the C terminus of E2F1 required to induce apoptosis, we also transferred various portions of the C terminus, including the marked-box and marked-box-adjacent region, into E2F3. Of these, the chimera containing the marked box and adjacent sequences (333113) was most active, exhibiting ≈50% of the wild-type E2F1 activity. In contrast, the reverse chimera (111331) was no better than E2F3. The fact that the 333113 chimera did not induce apoptosis as strongly as E2F1 suggests that the E2F1 transactivation domain is required for full activation of apoptosis (Fig. 4). Chimeras that contain combinations of two of the C-terminal domains, either the marked-box domain and the transactivation domain (111131) or the marked-box-adjacent sequence together with the transactivation domain (111311), were reduced but did retain some activity. Analysis of the additional chimeras containing only the marked-box domain (333133) or only the marked-box-adjacent sequence (333313) indicates that neither of these domains on their own was sufficient for conferring apoptotic activity. Taken together, these results suggest that sequences in the C-terminal region of E2F1 are necessary and sufficient for apoptotic activity and that there are likely contributions from multiple domains, with the marked-box and adjacent sequences being most critical.
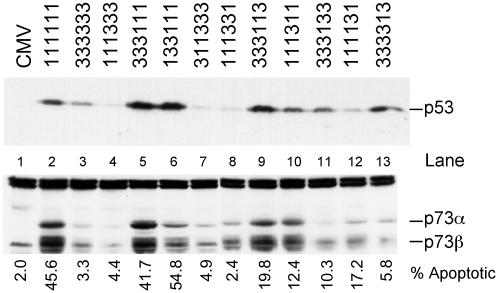
The E2F1 Apoptotic Domain Is Also Required for Induction of p53 and p73. To explore the potential mechanisms allowing these chimeras to induce apoptosis, we examined their ability to induce p53 and p73 protein levels. The induction of p53 accumulation likely involves an ability of E2F1 to induce p19ARF and then inhibit Mdm2-mediated degradation of p53 (14, 25). In contrast, the induction of p73 by E2F1 appears to be a direct effect on transcription of the p73 gene (27). Quiescent REF52 cells were infected with chimeric E2F adenoviruses at an moi of 75 and returned to serum starvation medium for 24 h. Whole cell extracts were prepared and analyzed by SDS/PAGE and Western blotting with p53 antiserum. As shown in Fig. 5, the ability of the chimeric proteins to induce p53 accumulation generally coincided with their capacity to induce apoptosis. In particular, E2F1 but not E2F3 induced p53 accumulation (lanes 2 and 3), and this induction was mirrored by the chimeras that most clearly defined apoptotic function. For instance, the chimera containing the E2F1 C terminus (333111) or the E2F1 marked-box and adjacent sequence (333113) were equal to wild-type E2F1 (lanes 5 and 9), whereas the reverse chimeras (111333 or 111331) were as defective as E2F3 (lanes 4 and 8). In general, the other chimeras containing only the E2F1 marked-box or adjacent region were reduced in their capacity to induce p53.
Fig. 5.
The E2F1 marked-box and adjacent domain are required for the induction of p53 and p73 protein accumulation. (Upper) Serum-starved REF52 cells were infected with the indicated E2F adenovirus (75 focus-forming units per cell) and returned to starvation medium for 24 h. Whole cell extracts were prepared and separated by SDS/PAGE. The levels of p53 protein were assessed by Western blotting. (Lower) We were unable to detect rat p73 with the available antiserum, so we used Saos-2 cells to assess expression of human p73 in response to the E2F chimeras. Saos-2 cells were plated at a density of 3 × 105 cells per 60-mm-diameter dish and grown in 10% serum for 24 h, then switched to 0.25% serum-containing medium for 48 h. Cells were infected with chimeric E2F-producing virus and were then returned to starvation medium. Nuclear extracts were prepared 24 h after infection, and 100 μg of protein was separated by SDS/PAGE and probed with anti-p73 antisera (Oncogene).
To assess p73 protein elevation in response to E2Fs, Saos-2 cells were serum-starved for 48 h, infected with the viruses producing chimeric E2Fs, and returned to starvation medium for 24 h. Nuclear extracts were prepared at 24 h, and equal amounts of protein were separated by SDS/PAGE and probed a p73 antiserum. The results of these assays were similar to the results observed with p53. Like the induction of p53, E2F1 but not E2F3 induced the expression of p73α and p73β (lanes 2 and 3). The chimeras containing the C-terminal sequences of E2F1 (333111 and 133111) were nearly equal to wild-type E2F1 (lanes 5 and 6). Likewise, the chimera containing the E2F1 marked-box domain and adjacent sequence (333113) was also capable of inducing p73, whereas the reverse chimera (111331) did not induce p73 (compare lanes 8 and 9). Chimeras containing only the E2F1 marked-box domain (333133) or only the adjacent sequence (333313) were not capable of inducing p73 (lanes 11 and 13), a result in contrast to the p53 result in which these chimeras retained some ability to induce p53. The chimera containing the marked-box-adjacent sequence as well as transactivation domain (111311) was nearly fully active in inducing p73.
Taken together, these observations support the view that induction of p53 and p73 coincides with the ability of E2F1 to induce apoptosis, dependent on the marked-box domain and adjacent sequence. The fact that there is not an exact quantitative relationship in each case between the ability to induce p53 or p73 and the ability to induce apoptosis suggests that the apoptotic function likely involves more than just the ability to induce p53 and p73.
Discussion
The Rb/E2F pathway occupies a central role in the critical cellular decisions that link proliferation and fate determination. E2Fs are required for the activation of a large group of genes encoding both DNA replication activities and other cell cycle regulatory activities; as such, E2F activities can be seen to be crucial for the transition of cells from G1 to S phase (1, 2). In addition, other E2Fs are clearly critical for the decisions associated with cell-cycle exit and terminal differentiation. Finally, it has also now become clear that the cellular processes controlling apoptosis also link to the Rb/E2F pathway. In one sense, this link can be seen as a mechanism to monitor the major proliferative pathway, serving as a checkpoint for abnormal events such as mutations in Rb or other activities that control E2F accumulation (42). Indeed, various studies now point to E2F1 as the critical effector of the apoptotic signal associated with deregulation of the Rb/E2F pathway. In another context, it serves to further add to the DNA-damage response as a result of the induction of E2F1 accumulation (23). Given the ability of E2F1 to serve as both a positive activator of cellular proliferation and an inducer of cell death, this E2F family member can be seen as a nexus in the decisions of cellular fate. The experiments we present here help to further delineate those cellular mechanisms responsible for controlling the E2F1 death signal as well as mechanisms underlying the specificity of E2F1 action in triggering apoptosis.
Pathways Controlling the Balance Between Proliferation and Death. The original observation that E2F1 triggered a p53-dependent apoptotic response provided a connection between the Rb/E2F cell proliferation pathway and the p53 cell fate pathway (9). Subsequent experiments provided a mechanistic basis for this connection by linking E2F1 to the activation of the p19ARF gene and control of Mdm2 and p53 accumulation (14, 25, 43, 44). This connection thus provides a basis for monitoring the normal activity of the Rb/E2F proliferation pathway; in a sense, the p19ARF/Mdm2/p53 connection can be seen as a checkpoint to monitor abnormal proliferative events (42). Nevertheless, it is also true that E2F1 accumulation takes place as part of the normal process of cellular proliferation; as such, it is obvious that the apoptotic function of E2F1 must be blocked in some fashion for cells to grow and survive. The work we present here provides evidence that the Ras–PI3K–Akt pathway plays such a role.
Many studies have documented the role of Ras and Ras-effector pathways in the normal process of cellular proliferation. Activation of Ras after growth stimulation leads to the initiation of a multitude of signaling events, some of which are clearly critical for initiating the action of the Rb/E2F pathway. For instance, Raf/MEK/Erk signaling is essential for activation of cyclin D transcription, resulting in the generation of cyclin D/cdk4 activity that then leads to Rb phosphorylation. Raf/MEK/Erk signaling is also important to allow the accumulation of Myc (45, 46), and various observations suggest a role for Myc in enhancing the accumulation of E2Fs (45, 47). Although other work has provided evidence of the Ras–Raf–MEK–Erk pathway in mediating cell survival, the experiments we present here demonstrate that this Ras effector arm is not involved in suppressing the E2F1 apoptotic signal, but rather it is the PI3K/Akt Ras effector that appears to be essential for blocking the E2F1 signal. Although the precise mechanism by which Akt blocks the E2F1 signal remains to be elucidated, it is clear from our studies that it prevents the induction of p53 accumulation. One mechanism for this action could involve an Akt-mediated phosphorylation of Mdm2, facilitating nuclear localization of Mdm2 and degradation of p53 (38, 48).
A Mechanistic Basis for Specificity of E2F1-Induced Apoptosis. The capacity of E2F1 to specifically induce apoptosis coincides with previous work that has shown an ability of E2F1 to preferentially induce the expression of several genes involved in apoptotic pathways. This includes the activation of the p19ARF gene (14, 25), leading to the accumulation of p53 caused by p19ARF-mediated inhibition of the Mdm2 ubiquitin ligase. Moreover, other work has provided evidence of an E2F1-mediated induction of various other genes involved in apoptosis, such as Apaf1 (26), caspases 3, 7, 8, and 9 (49), and p73 (27). Thus, it seems plausible that the specificity in E2F1-mediated induction of apoptosis would involve an ability to specifically induce the transcription of genes involved in the apoptotic process. The work we present here connects the specificity of apoptosis mediated by the E2F1 marked-box domain with the role of this domain in the induction of apoptotic activities.
Importantly, other recent work has implicated the marked-box domain in protein interactions that dictate specificity of promoter recognition, following from the initial observation that the ability of an adenovirus E4 protein to target E2F to the viral E2 promoter depended on an interaction with the marked-box domain (50). In particular, several proteins involved in transcription control have been identified that specifically associate with individual E2F family members and that do so dependent on the marked-box domain. For example, E2F2 and E2F3 have been shown to interact with the RYBP protein, which also provides an interaction with the YY1 transcription factor (51). In addition, E2F3 has also been shown to interact with the E box transcription factor TFE3, and again, this depends on the marked-box domain (52). In both instances, these interactions coincide with the ability of the proteins to synergistically activate transcription of target genes that contain E2F sites and either YY1 elements or E box elements. Importantly, these interactions provide a basis for specificity of transcription control within the E2F family because the interactions are restricted to particular E2F family members. For instance, although E2F3 interacts with TFE3 to facilitate formation of a promoter complex and activation of transcription, E2F1 does not interact with TFE3, nor does it associate with those promoters containing E box elements. Although a specific partner for E2F1 has not yet been identified, it is tempting to speculate that the basis for E2F1 specificity in the induction of apoptosis and activation of genes important for apoptosis reflects an ability to function with another transcription factor that, together with E2F1, provides specificity for the relevant target genes.
Acknowledgments
We thank Kaye Culler for assistance in the preparation of the manuscript. T.C.H. was supported by American Cancer Society Postdoctoral Fellowship PF-02-004-01-CCG. J.R.N. is an Investigator with the Howard Hughes Medical Institute.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: moi, multiplicity of infection; HA, hemagglutinin; PI3K, phosphoinositide 3-kinase; CMV, cytomegalovirus; MEK, mitogen-activated protein kinase kinase.
References
- 1.Dyson, N. (1998) Genes Dev. 12, 2245–2262. [DOI] [PubMed] [Google Scholar]
- 2.Nevins, J. R. (1998) Cell Growth Differ. 9, 585–593. [PubMed] [Google Scholar]
- 3.Leone, G., DeGregori, J., Yan, Z., Jakoi, L., Ishida, S., Williams, R. S. & Nevins, J. R. (1998) Genes Dev. 12, 2120–2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Humbert, P. O., Verona, R., Trimarchi, J. M., Rogers, C., Dandapani, S. & Lees, J. A. (2000) Genes Dev. 14, 690–703. [PMC free article] [PubMed] [Google Scholar]
- 5.Humbert, P. O., Rogers, C., Ganiatsas, S., Landsberg, R. L., Trimarchi, J. M., Dandapani, S., Brugnara, C., Erdman, S., Schrenzel, M., Bronson, R. T., et al. (2000) Mol. Cell 6, 281–291. [DOI] [PubMed] [Google Scholar]
- 6.Rempel, R. E., Saenz-Robles, M. T., Storms, R., Morham, S., Ishida, S., Engel, A., Jakoi, L., Melhem, M. F., Pipas, J. M., Smith, C., et al. (2000) Mol. Cell 6, 293–306. [DOI] [PubMed] [Google Scholar]
- 7.Johnson, D. G., Schwarz, J. K., Cress, W. D. & Nevins, J. R. (1993) Nature 365, 349–352. [DOI] [PubMed] [Google Scholar]
- 8.Wu, L., Timmers, C., Maiti, B., Saavedra, H. I., Sang, L., Chong, G. T., Nuckolls, F., Giangrande, P., Wright, F. A., Field, S. J., et al. (2001) Nature 414, 457–462. [DOI] [PubMed] [Google Scholar]
- 9.Wu, X. & Levine, A. J. (1994) Proc. Natl. Acad. Sci. USA 91, 3602–3606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kowalik, T. F., DeGregori, J., Schwarz, J. K. & Nevins, J. R. (1995) J. Virol. 69, 2491–2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kowalik, T. F., DeGregori, J., Leone, G. & Nevins, J. R. (1998) Cell Growth Differ. 9, 113–118. [PubMed] [Google Scholar]
- 12.Qin, X.-Q., Livingston, D. M., Kaelin, W. G. & Adams, P. D. (1994) Proc. Natl. Acad. Sci. USA 91, 10918–10922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shan, B. & Lee, W.-H. (1994) Mol. Cell. Biol. 14, 8166–8173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DeGregori, J., Leone, G., Miron, A., Jakoi, L. & Nevins, J. R. (1997) Proc. Natl. Acad. Sci. USA 94, 7245–7250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Field, S. J., Tsai, F.-Y., Kuo, F., Zubiaga, A. M., Kaelin, W. G., Jr., Livingston, D. M., Orkin, S. H. & Greenberg, M. E. (1996) Cell 85, 549–561. [DOI] [PubMed] [Google Scholar]
- 16.Zhu, J. W., DeRyckere, D., Li, F. X., Wan, Y. Y. & DeGregori, J. (1999) Cell Growth Differ. 10, 829–838. [PubMed] [Google Scholar]
- 17.Jacks, T., Fazeli, A., Schmitt, E. M., Bronson, R. T., Goodell, M. A. & Weinberg, R. A. (1992) Nature 359, 295–300. [DOI] [PubMed] [Google Scholar]
- 18.Lee, E. Y. H. P., Chang, C.-Y., Hu, N., Wang, Y.-C. J., Lai, C.-C., Herrup, K., Lee, W.-H. & Bradley, A. (1992) Nature 359, 288–294. [DOI] [PubMed] [Google Scholar]
- 19.Pan, H., Yin, C., Dyson, N. J., Harlow, E., Yamasaki, L. & van Dyke, T. (1998) Mol. Cell 2, 283–292. [DOI] [PubMed] [Google Scholar]
- 20.Tsai, K. Y., Hu, Y., Macleod, K. F., Crowley, D., Yamasaki, L. & Jacks, T. (1998) Mol. Cell 2, 293–304. [DOI] [PubMed] [Google Scholar]
- 21.Yamasaki, L., Bronson, R., Williams, B. O., Dyson, N. J., Harlow, E. & Jacks, T. (1998) Nat. Genet. 18, 360–364. [DOI] [PubMed] [Google Scholar]
- 22.Ziebold, U., Reza, T., Caron, A. & Lees, J. A. (2001) Genes Dev. 15, 386–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin, W.-C., Lin, F.-T. & Nevins, J. R. (2001) Genes Dev. 15, 1833–1845. [PMC free article] [PubMed] [Google Scholar]
- 24.Honda, R., Tanaka, H. & Yasuda, H. (1997) FEBS Lett. 420, 25–27. [DOI] [PubMed] [Google Scholar]
- 25.Bates, S., Phillips, A. C., Clark, P. A., Stott, F., Peters, G., Ludwig, R. L. & Vousden, K. H. (1998) Nature 395, 124–125. [DOI] [PubMed] [Google Scholar]
- 26.Moroni, M. C., Hickman, E. S., Denchi, E. L., Caprara, G., Colli, E., Cecconi, F., Muller, H. & Helin, K. (2001) Nat. Cell Biol. 3, 552–558. [DOI] [PubMed] [Google Scholar]
- 27.Irwin, M., Martin, M. C., Phillips, A. C., Seelan, R. S., Smith, D. I., Liu, W., Flores, E. R., Tsai, K. Y., Jacks, T., Vousden, K. H., et al. (2000) Nature 407, 645–648. [DOI] [PubMed] [Google Scholar]
- 28.Croxton, R., Ma, Y., Song, L., Haura, E. B. & Cress, W. D. (2002) Oncogene 21, 1359–1369. [DOI] [PubMed] [Google Scholar]
- 29.Phillips, A. C., Ernst, M. K., Bates, S., Rice, N. R. & Vousden, K. H. (1999) Mol. Cell 4, 771–781. [DOI] [PubMed] [Google Scholar]
- 30.He, T.-C., Zhou, S., da Costa, L. T., Yu, J., Kinzler, K. W. & Vogelstein, B. (1998) Proc. Natl. Acad. Sci. USA 95, 2509–2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cook, J. G., Park, C. H., Burke, T. W., Leone, G., DeGregori, J., Engel, A. & Nevins, J. R. (2002) Proc. Natl. Acad. Sci. USA 99, 1347–1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Muller, H., Bracken, A. P., Vernell, R., Moroni, M. C., Christians, F., Grassilli, E., Prosperini, E., Vigo, E., Oliner, J. D. & Helin, K. (2001) Genes Dev. 15, 267–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ries, S., Biederer, C., Woods, D., Shifman, O., Shirasawa, S., Sasazuki, T., McMahon, M., Oren, M. & McCormick, F. (2000) Cell 103, 321–330. [DOI] [PubMed] [Google Scholar]
- 34.Brazil, D. P. & Hemmings, B. A. (2001) Trends Biochem. Sci. 26, 657–664. [DOI] [PubMed] [Google Scholar]
- 35.Ozes, O. N., Mayo, L. D., Gustin, J. A., Pfeffer, S. R., Pfeffer, L. M. & Donner, D. B. (1999) Nature 401, 82–85. [DOI] [PubMed] [Google Scholar]
- 36.Staal, S. P. & Hartley, J. W. (1988) J. Exp. Med. 167, 1259–1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bellacosa, A., Testa, J. R., Staal, S. P. & Tsichlis, P. N. (1991) Science 254, 274–277. [DOI] [PubMed] [Google Scholar]
- 38.Mayo, L. D. & Donner, D. B. (2001) Proc. Natl. Acad. Sci. USA 98, 11598–11603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou, B. P., Liao, Y., Xia, W., Zou, Y., Spohn, B. & Hung, M.-C. (2001) Nat. Cell Biol. 3, 973–982. [DOI] [PubMed] [Google Scholar]
- 40.Ashcroft, M., Ludwig, R. L., Woods, D. B., Copeland, T. D., Weber, H. O., MacRae, E. J. & Vousden, K. H. (2002) Oncogene 21, 1955–1962. [DOI] [PubMed] [Google Scholar]
- 41.Gottlieb, T. M., Leal, J. F., Seger, R., Taya, Y. & Oren, M. (2002) Oncogene 21, 1299–1303. [DOI] [PubMed] [Google Scholar]
- 42.Sherr, C. J. (1998) Genes Dev. 12, 2984–2991. [DOI] [PubMed] [Google Scholar]
- 43.Kamijo, T., Weber, J. D., Zambetti, G., Zindy, F., Roussel, M. F. & Sherr, C. J. (1998) Proc. Natl. Acad. Sci. USA 95, 8292–8297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang, Y., Xiong, Y. & Yarbrough, W. G. (1998) Cell 92, 725–734. [DOI] [PubMed] [Google Scholar]
- 45.Sears, R., Nuckolls, F., Haura, E., Taya, Y., Tamai, K. & Nevins, J. R. (2000) Genes Dev. 14, 2501–2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sears, R., Leone, G., DeGregori, J. & Nevins, J. R. (1999) Mol. Cell 3, 169–179. [DOI] [PubMed] [Google Scholar]
- 47.Sears, R., Ohtani, K. & Nevins, J. R. (1997) Mol. Cell. Biol. 17, 5227–5235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ogawara, Y., Kishishita, S., Obata, T., Isazawa, Y., Suzuki, T., Tanaka, K., Masuyama, N. & Gotoh, Y. (2002) J. Biol. Chem. 277, 21843–21850. [DOI] [PubMed] [Google Scholar]
- 49.Nahle, Z., Polakoff, J., Davuluri, R. V., McCurrach, M. E., Jacobson, M. D., Narita, M., Zhang, M. Q., Lazebnik, Y., Bar-Sagi, D. & Lowe, S. W. (2002) Nat. Cell Biol. 4, 859–864. [DOI] [PubMed] [Google Scholar]
- 50.Jost, C. A., Ginsbreg, D. & Kaelin, W. G., Jr. (1996) Virology 220, 78–90. [DOI] [PubMed] [Google Scholar]
- 51.Schlisio, S., Halperin, T., Vidal, M. & Nevins, J. R. (2002) EMBO J. 21, 5775–5786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Giangrande, P. H., Hallstrom, T. C., Tunyaplin, C., Calame, K. & Nevins, J. R. (2003) Mol. Cell. Biol. 23, 3707–3720. [DOI] [PMC free article] [PubMed] [Google Scholar]