Abstract
Yeast strains with mutations in both TEL1 and MEC1 have short telomeres and elevated rates of chromosome deletions. By using a PCR assay, we demonstrate that mec1 tel1 strains also have telomere–telomere fusions (T-TFs). T-TFs require Lig4p (a ligase required for nonhomologous end-joining DNA repair). The highest rates of T-TFs are found in strains with combination of mutations that affect telomere length and DNA damage checkpoints (mec1 tel1, mec3 tel1, mre11 mec1, and ddc1 tel1 strains). Examining many mutant genotypes, we find good agreement between the level of T-TFs and the rate of chromosomal deletions. In addition, if telomeres are elongated in a mec1 tel1 strain, we eliminate T-TFs and reduce the deletion rate. The correlation between the level of T-TFs and the rate of deletions argues that many of these deletions reflect a cycle of T-TF formation (resulting in dicentric chromosomes), followed by chromosome breakage.
In eukaryotes, loss of telomeric sequences is often associated with the generation of chromosome aberrations. Increased frequencies of end-to-end chromosome associations, suggestive of telomere fusions, have been observed in mammalian cells with mutations in ATM, Ku, DNA-PK, and TRF2 (1, 2). In addition, chromosomes in transformed mammalian cells in crisis have short telomeres and high frequencies of end-to-end associations; after activation of telomerase, there is a reduction in the frequency of chromosome aberrations (3).
The connection between short telomeres and chromosome aberrations has also been examined in Saccharomyces cerevisiae. Yeast strains with mutations in both TEL1 (an ATM homologue) and MEC1 (an ATR homologue) have short telomeres and undergo cellular senescence (4, 5). These strains also have high rates of aberrant chromosomes, including translocations, dicentrics, and circular chromosomes (6, 7). Elevated genetic instability is observed in est1 yeast strains, associated with fusions of subtelomeric sequences (8), and circular chromosomes (suggestive of telomere fusions) are observed in est2 nej1 strains (9). Fusions between telomeric sequences and HO-induced DNA breaks occur at a frequency of >1/1,000/HO-induced end in tel1 tlc1 strains (10).
In our previous study of genome instability in mec1 tel1 strains (7), we selected for derivatives that had deleted the CAN1 gene. Most of these deletions were nonreciprocal translocations and, in 40% of these translocations, telomeric or subtelomeric sequences were located at the translocation breakpoint. These results suggested that mec1 tel1 strains might have a high rate of fusions involving two telomeric sequences. Below, we demonstrate the existence of telomere-telomere fusions (T-TFs) and provide evidence that these fusions generate other types of chromosome aberrations.
Materials and Methods
Strain Construction. All strains in this study were isogenic with W303a (11) except for changes introduced by transformation or by crosses with isogenic strains. The construction of the strains and sequences of oligonucleotides used in this study are described in Tables 3 and 4, which are published as supporting information on the PNAS web site, www.pnas.org.
PCR Assay to Detect T-TFs. T-TFs were detected by using a PCR assay. One primer (ChrXV-UP; 5′-AAGAATTCTATGGTTAAATGGGGCAGGGTAACG) had DNA sequences from the X element of chromosome XVL (coordinates 183–207; Stanford Genome Database) and the second (ChrV-30; 5′-AAGAATTCGGTA AGAGACA ACAGGGCT TGGAGG) had sequences from the Y′ element of chromosome VR (coordinates 576754–576778). To control for the efficiency of the PCR reaction in different DNA samples, we used primers (HIS4-UP and HIS4-DN) that amplified the nontelomeric HIS4 sequences. Details of the PCR conditions are in Supporting Methods, which is published as supporting information on the PNAS web site.
Procedures for Measuring the Rates of CAN1 Deletions and for Performing Chromatin Immunoprecipitation (ChIP) Assays. Forward mutation rates at the CAN1 locus were measured in haploid strains by using standard methods (12). For each strain, we determined the fraction of the can1 mutations that were deletions by PCR by using primers CAN-LW1 and CAN-UP1 (Table 4). Strains that retain the CAN1 gene produce a PCR product of 300 bp. The methods for the ChIP are described in Supporting Methods.
Results
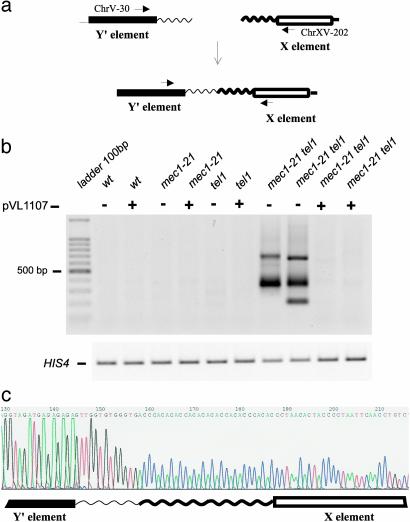
T-TFs and Genome Stability in mec1 tel1 Cells. Strains with the mec1 tel1 genotype undergo cellular senescence, followed by formation of “survivors” similar to those observed for strains lacking telomerase (4). Haploid mec1 tel1 strains were made by sporulating doubly heterozygous diploids; the haploid-viable mec1-21 allele (13) was used. To determine whether these strains had T-TFs, we used a PCR assay. One primer contained sequences from the subtelomeric X element located ≈60 bp from the junction with the terminal poly G1–3T sequences. The other had sequences of the Y′ element located ≈150 bp from the junction with the poly G1–3T tract. A fusion between the poly G1–3T sequences of a chromosome with a terminal X repeat with the telomeric repeats of a chromosome with a Y′ repeat would generate a PCR product of 210 bp plus the amount of DNA in telomeric tracts (Fig. 1a). All DNA samples were also analyzed by using primers that amplified the nontelomeric HIS4 gene.
Fig. 1.
Analysis of T-TFs by PCR. (a) T-TF detection system. By performing PCR with one primer derived from the Y′ subtelomeric repeat and one from the X subtelomeric repeats, we detect a subset of T-TFs. (b Upper) T-TF assays of DNA isolated from wild-type, mec1–21, tel1, and mec1–21 tel1 strains with (+) and without (–) a plasmid (pVL1107) that elongates telomeres by a Mec1- and Tel1-independent mechanism. Strain names in lanes 2–11 (lane 1 containing the marker DNA) are: JMY309-1a, PM184-3d, JMY309-1b, PM184-2b, JMY309-1c, PM184-7c, JMY309-1d, PM184-1d, and PM184-12a (lanes 10 and 11). (Lower) A control PCR (labeled HIS4). Strain names and associated genotypes are given in Supporting Methods.(c) DNA sequence of a T-TF. A PCR fragment of the size diagnostic of a T-TF derived from a mec1–21 tel1 strain was cloned and sequenced. The DNA sequence shows a fusion between the poly G1–3T tract on the Y′ repeat and the poly C1–3A tract of an X repeat.
Of 27 independent mec1 tel1 spore cultures tested, all had one or more abundant PCR product (indicated by ++ in Table 1) of the sizes expected for T-TFs (Fig. 1b). Abundant fragments of this type were not observed in haploids of the tel1, mec1, or wild-type genotypes (Table 1 and Fig. 1b). In Fig. 1b, the relevant samples are those that have a minus sign above the gel lane, indicating that they lack the plasmid pVL1107 (discussed below). Although most of the PCR products were 300–400 bp in size, some samples had PCR fragments >500 bp; in Table 1, the numbers of samples of this type are shown in parentheses.
Table 1. PCR analysis of T-TFs.
| Abundance of PCR fragment diagnostic of T-TF†
|
Real-time PCR analysis‡
|
||||
|---|---|---|---|---|---|
| Genotype* | ++ | + | ± | - | No. of T-TFs per genome |
| Wild type | (4) | 21 | 1.9 × 10-7 (1-3.4) | ||
| te/1 | 1 (2) | 15 | 1 × 10-7 (0.4-3.9) | ||
| mec1-21 | 2 | 4 | 14 | 5.9 × 10-7 (2-18) | |
| mec1-21 te/l | 27 | 1.1 × 10-5 (0.8-2.2) | |||
| mec1-21 mre11 | 4 | 6 | 3 | 2 | ND |
| mec1-21 te/1 mre/1 | 7 | 3 | ND | ||
| te/1 mre11 | 2 | 6 | ND | ||
| mec1-21 te/1 + pVL1107 | 1 | 6 | 1.3 × 10-6 (0.6-2.5) | ||
| t/c1 | 1 | 2 | 8 | 2.2 × 10-6 (0.4-6.6) | |
| mec1-21 t/c1 | 3 | 10 | 2 | 1 | 2.5 × 10-6 (0.7-18) |
| mec3 te/1 | 7 | 4 | 8 × 10-5 (3.5-16) | ||
| ddc1 te/1 | 7 | 3 | 2.3 × 10-5 (1.6-5.3) | ||
| pds1 te/1 | 2 | 2 | 8 | ND | |
| yku70 | 4 | ND | |||
| yku80 | (1) | (6) | (3) | ND | |
| mec1-21 yku70 | 4 | ND | |||
| mec1-21 yku80 | (1) | (4) | 5 | ND | |
| te/1 yku70 | 4 | ND | |||
| te/1 yku80 | (4) | 5 | ND | ||
| mec1-21 te/1 yku70 | (2) | (3) | (1) | 5.8 × 10-6 (0.4-23) | |
| mec1-21 te/1 yku80 | (1) | (12) | 4.4 × 10-6 (3.3-11) | ||
| mec1-21 te/1 lig4 | 1 | 10 | 1.3 × 10-6 (0.4-4.1) | ||
| mec1-21 mre11 lig4 | (3) | 8 | ND | ||
DNA was isolated from haploid strains of the indicated genotypes. Most of these haploids were derived from sporulating heterozygous diploids and subcloned three times before DNA isolation. We examined multiple strains with the same genotypes (strain names are given in Supporting Methods). We performed PCR amplifications of these samples with primers diagnostic of TT-Fs. ND, not done.
Only relevant differences from the progenitor genotype of W303a (a leu2-3,112 his3-11 ura3-1 ade2-1 trp1-1 can1-100 rad5-535) are shown in the “Genotype” column.
The numbers in each column indicate the number of DNA samples with PCR products that were abundant (++), moderately abundant (+), weak (±), or absent (-). Numbers without parentheses show the number of samples with a PCR fragment of ≈350 bp, and numbers with parentheses indicate the number of samples resulting in a PCR product of ≈500 bp or greater.
Real-time PCR was performed by using two sets of primers, a T-TF-specific pair and a pair that amplified single-copy sequences derived from HIS4. By calculating the differences in the number of PCR cycles required to yield similar amounts of product during the exponential part of the amplification process, we determined the relative frequency of T-TFs and the single-copy sequence in each strain. Reactions were done on three to seven strains of each genotype. Numbers in parentheses are 95% confidence intervals.
We cloned and sequenced the T-TF PCR products from 21 independent mec1 tel1 strains, finding three classes: Class 1 (57%), fusions between two terminal simple repetitive tracts; Class 2 (14%), fusions of internal sequences of the X repeat with a telomeric tract of the Y; Class 3 (29%), fusions between internal Y′ sequences and the telomeric repeat of an X telomere. Class 1 fusions are imperfect palindromes with a poly G1–3T sequence joined to a poly C1–3A sequence (Fig. 1c). Because of the difficulty in sequencing palindromic insertions, we were unable to sequence the junctions between the telomeric repeats in any event in which there were >100 bp of telomeric DNA. One potential artifact in using PCR to detect T-TFs in the wild-type and mec1 strains is that a PCR fragment containing a fusion between two wild-type length telomeres might be more difficult to amplify than a fusion between short telomeric repeats. We showed in a control experiment (see Supporting Methods) that fusions between telomeric repeats of wild-type length were readily detected by PCR.
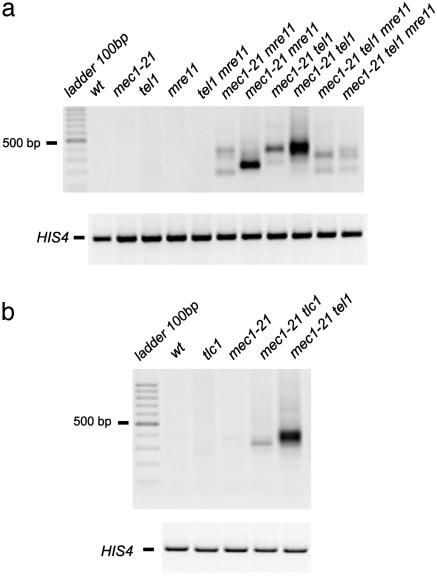
Strains with mutations in MRE11, RAD50,or XRS2 have short telomeres (14, 15), and the MRX proteins function in the same pathway of telomere length regulation as Tel1p (16). Consistent with this conclusion, we found that both mec1 mre11 and mec1 tel1 mre11 strains had substantial levels of T-TFs in most DNA samples (Table 1 and Fig. 2a). Strains with the tel1 mre11 genotype did not efficiently form T-TFs (Table 1 and Fig. 2a), as expected, because Tel1p and Mre11p are epistatic in the regulation of telomere length.
Fig. 2.
T-TF assay of DNA isolated from yeast strains with mutations in MRE11 or TLC1.(a) Effect of the mre11 mutation on T-TFs in combination with the tel1 and mec1 mutations. PCRs the same as those described in Fig. 1 were done. Strain names in lanes 2–12 are: PM198-2b, PM198-3d, PM198-13d, PM198-4b, PM198-6a, PM198-6b (lanes 7 and 8), PM198-8d (lanes 9 and 10), and PM198-12c (lanes 11 and 12). (b) Effect of the tlc1 mutation on T-TFs with and without the additional mec1–21 mutation. Strain names in lanes 2–6 are: KRY237-9a, KRY237-9d, KRY237-9c, KRY237-9b, and KRY237-8d.
In addition to the semiquantitative PCR analysis in which we visually compared the amount of T-TF-specific and control PCR fragments, we also did real-time PCR of samples derived from some genotypes. For each DNA sample, we did two PCR reactions, one to amplify the single-copy HIS4 control sequences and one to amplify T-TFs. By comparing the number of PCR cycles required to produce the same amount of the two PCR fragments, we calculated the relative amounts of these two DNA sequences in the DNA sample; control experiments indicate that the two different fragments are amplified with equal facility (see details in Supporting Methods). As expected from our previous PCR analysis, T-TFs in wild-type, tel1, and mec1 strains are found at very low frequencies (1–6 × 10–7 per genome), whereas the mec1 tel1 strain had a frequency of ≈10–5 per genome (Table 1). Our analysis is restricted to T-TFs that involve one Y′-bearing telomere and one X-bearing telomere. From an analysis of telomeres in the sequenced yeast genome by E. Louis (www.le.ac.uk/ge/ejl12), we estimate that we detect about half of the total T-TFs.
We and others previously showed that mec1 tel1 strains had high rates of genome instability (7, 17). One method of assaying genome instability is by measuring the rate of can1 (canavanine-resistant) mutations and multiplying that rate by the fraction of can1 mutations that are deletions (determined by PCR analysis). We showed previously that most can1 deletions reflect loss of the CAN1 gene and all centromere-distal sequences, as a consequence of a nonreciprocal translocation. As shown in Table 2, the rate of such deletions is much higher in mec1 tel1 strains than in wild-type or single mutant strains.
Table 2.
Rates of forward mutation and deletion formation at the CAN1 locus in strains with mutations affecting genome stability
| Genotype | Mutation rate (95% confidence limits)† | Rate of deletions‡ |
|---|---|---|
| Wild-type* | 2.4 × 10-7 (1.8-3.6 × 10-7) | <1 × 10-9 |
| mec1-21* | 3.1 × 10-7 (2.5-3.9 × 10-7) | 9.5 × 10-8 |
| te/1* | 3.4 × 10-7 (2.7-4.8 × 10-7) | 5.1 × 10-8 |
| mec1-21 te/1 | 1.0 × 10-5 (0.6-1.4 × 10-5) | 4.6 × 10-6 |
| mec1-21 te/1 + pVL1107 | 8 × 10-7 (2.7-4.8 × 10-7) | 2.4 × 10-7 |
| t/c1* | 2.0 × 10-6 (0.7-4.5 × 10-6) | 5.6 × 10-8 |
| t/c1§ | 1.3 × 10-6 (1.2-2 × 10-6) | 4 × 10-7 |
| mec1-21 t/c1 | 2.8 × 10-6 (1.8-4.7 × 10-6) | 3.1 × 10-7 |
| mec3 te/1 | 9.0 × 10-6 (3.2-16 × 10-6) | 5.4 × 10-6 |
| ddc1 te/1 | 7.5 × 10-6 (3.2-12 × 10-6) | 3.5 × 10-6 |
| pds1 te/1 | 5.2 × 10-6 (2.8-13 × 10-6) | 3.3 × 10-7 |
| mec1-21 lig4 | 2.5 × 10-7 (1.5-3.2 × 10-7) | <1 × 10-9 |
| te/1 lig4 | 3.6 × 10-7 (3.1-4.4 × 10-7) | 1.8 × 10-8 |
| mec1-21 te/1 lig4 | 1.8 × 10-6 (0.8-5.7 × 10-6) | 4.4 × 10-7 |
| te/1 yku80 | 4 × 10-7 (3.6-7 × 10-7) | 2 × 10-8 |
| mec1-21 yku80 | 5 × 10-7 (3.6-5.7 × 10-7) | 2.5 × 10-8 |
| mec1-21 te/1 yku80 | 1.5 × 10-6 (0.8-2.9 × 10-6) | 5.3 × 10-7 |
| te/1 yku70 | 8.6 × 10-7 (4.9-12.3 × 10-7) | <1 × 10-9 |
| mec1-21 yku70 | 4.1 × 10-7 (2.8-6.3 × 10-7) | 2.1 × 10-8 |
| mec1-21 te/1 yku70 | 1.4 × 10-6 (0.6-3.2 × 10-6) | 2.8 × 10-7 |
Table 5, which is published as supporting information on the PNAS web site, is an extended form of this table.
Data from ref. 7.
For each strain, we measured the frequency of canavanine-resistant cells in 20 independent cultures. Rate calculations were done as described (12).
The rate of deletions was calculated by multiplying the forward rate of mutations at the CAN1 locus by the fraction of these mutations that were deletions (determined by PCR analysis of 20 independent can1 mutants).
The rate of deletions for this strain was calculated by a method different from that used for the other strains (details are in Table 5).
Suppression of T-TF Formation and Genome Instability of mec1 tel1 Strains by Restoring Wild-Type Length Telomeres. The high rate of T-TFs in mec1 tel1 strains could reflect the telomere defect in this strain, the DNA damage checkpoint deficiency, or both factors. To distinguish among these possibilities, we constructed derivatives of the mec1 tel1 strains in which the telomere defect, but not the DNA damage checkpoint deficiency, was alleviated. Tsukamoto et al. (18) showed that expression of the Cdc13p-Est2p fusion protein (19) suppresses the short telomere phenotype of mec1 tel1 strains. We transformed wild-type, tel1, mec1, and mec1 tel1 strains with a plasmid (pVL1107) encoding the fusion protein and showed, by Southern analysis, that telomere lengths in all of the transformed strains were of wild-type length (data not shown). In the mec1 tel1 strain with pVL1107, T-TFs were largely suppressed as assayed by both PCR procedures (Table 1; Fig. 1b). In addition, the CAN1 deletion rate in the mec1 tel1 strain with the plasmid was ≈20-fold less than the strain lacking the plasmid (Table 2). The plasmid had no significant effect on the rate of can1 mutations in the wild-type, tel1, or mec1 strains (data not shown).
The cosuppression of T-TFs and genome instability by the pVL1107 plasmid argues that the telomere defect of mec1 tel1 strains is an important factor in the genetic instability. The longer telomeres resulting from expression of the Cdc13-Est2 fusion protein may recruit telomere-binding proteins resulting in a telomere “cap” or the fusion protein itself may form the “cap.” We used real-time PCR to examine T-TFs in the mec1 tel1 strain immediately after loss of pVL1107 and found a level of T-TFs (5.8 × 10–6 per genome) close to that observed in the mec1 tel1 strain that had never had the plasmid (1.1 × 10–5 per genome). Because most of the telomeres in the strain that lost the plasmid were very short, however, this observation does not distinguish between the two models.
T-TFs in Other Strains with Mutations Affecting Telomere Length and/or the DNA Damage Checkpoints. Because the highest frequency of T-TFs was observed in the mec1 tel1 strain, which had mutations affecting both telomere length and the DNA damage checkpoint, we examined strains with other mutations in these two pathways. We detected T-TFs in tlc1 strains (telomerase-negative), although the level was less than observed in mec1 tel1 strains (Fig. 2b). Different tlc1 derivatives had somewhat different T-TF frequencies, and the effect of the tlc1 mutation on the rate of CAN1 deletions was somewhat variable. We also examined a tlc1 mec1 strain. The double mutant had more T-TFs than the tlc1 strain in the semiquantitative PCR assay, but no significant difference was observed by the real-time PCR assay. In addition, the rate of CAN1 deletions in the tlc1 mec1 strain was similar to that observed in one of the tlc1 strains (Table 2). We cloned and sequenced the T-TF PCR fragments from three independent tlc1 isolates: one involved X and Y′ poly G1–3T repeats, and two were fusions of Y′ telomeric repeats to nontelomeric X sequences. We also analyzed two PCR fragments derived from the tlc1 mec1 strain: one involved X and Y′ simple repeats, and one was a fusion of X telomeric repeats to Y′ nontelomeric sequences.
Strains with the mec3 tel1, ddc1 tel1, and pds1 tel1 genotypes have rates of deletions and other chromosome rearrangements similar to those observed in mec1 tel1 strains (6, 17). Mec3p and Ddc1p affect early steps in the DNA damage checkpoint, whereas Pds1p affects a late step (20). In most of our experiments, we examined strains for T-TFs after three subclonings (≈75 divisions). Although no T-TFs were observed in any of the three mutants after three subclonings, we found substantial levels of T-TFs in the mec3 tel1 and ddc1 tel1 strains and low levels of T-TFs in the pds1 tel1 strain, after seven subclonings (Fig. 3a, Table 1); we found very low levels of T-TFs in the mec3 and ddc1 single mutant strains (2.2 × 10–7 and 0.8 × 10–7 per genome, respectively). The CAN1 deletion rates in mec3 tel1 and ddc1 tel1 strains were very high, similar to those observed in the mec1 tel1 strain (Table 2). The rate of CAN1 deletions in the pds1 tel1 strain was ≈10-fold lower, consistent with the lower frequency of T-TFs.
Fig. 3.
Analysis of T-TFs in strains with mutations affecting DNA damage checkpoints (ddc1, mec3, and pds1) or nonhomologous DNA end-joining (yku70, yku80, and lig4). (a) Effect of mutations in pds1, ddc1, mec3, yku70, and yku80 on T-TF formation. Strains of the ddc1 tel1, mec3 tel1, pds1 tel1,or mec1 tel1 genotypes have the 350-bp PCR fragment diagnostic of T-TFs involving short telomeres, whereas mec1–21 tel1 yku70 and mec1–21 tel1 yku80 strains have multiple larger PCR fragments diagnostic of T-TFs involving longer telomeres. Strain names in lanes 2–12 are: PM197-1a, PM208 (lanes 3 and 4), PM197-7b, PM203-17c, PM195-11d, PM207 (lanes 8 and 9), PM206 (lanes 10 and 11), and PM197-8d. (b) Effect of the lig4 mutation on T-TF formation in association with tel1 and/or mec1 mutations. Strain names in lanes 1–10 are: PM183-1a, PM183-2c, PM183-6b, PM183-2a, PM183-1b (lanes 5 and 6), PM183-3d, PM183-12a, and PM183-13c (lanes 9 and 10).
Roles of the Nonhomologous End-Joining (NHEJ) Proteins (Lig4p and the Ku Proteins) in T-TFs. T-TF formation resembles NHEJ events previously described in S. cerevisiae. Efficient NHEJ requires the Mre11, Rad50, Xrs2, Yku70, Yku80, Lig4, Lif1, and Nej1 proteins (21, 22). Because we observe T-TFs in mec1 mre11 strains, the formation of T-TFs does not depend on Mre11p. Fusions between an HO-induced DNA break and telomeric sequences depend on Lig4p (an NHEJ-specific ligase), as is the formation of circular yeast chromosomes (9, 10). Because strains of the mec1 tel1 lig4 and the mec1 mre11 lig4 genotypes formed T-TFs inefficiently (Fig. 3b), Lig4p is required for efficient T-TF formation. We also measured the rate of CAN1 deletions in mec1 lig4, tel1 lig4, and mec1 tel1 lig4 strains (Table 2). The deletion rate in the mec1 tel1 lig4 triple mutant was ≈10-fold less than that observed in the mec1 tel1 strain. The reduced T-TFs correlated with reduced genome instability suggest that much (although not all) of the genome instability in the mec1 tel1 strain is a consequence of T-TF formation.
We also examined T-TF formation in yku70, yku80, mec1 yku70, mec1 yku80, tel1 yku70, tel1 yku80, mec1 tel1 yku70, and mec1 tel1 yku80 strains. Most spores with the triple mutant genotype failed to give rise to a colony, and the surviving strains with this genotype were very slow-growing. The 350-bp fragment diagnostic of T-TFs involving short telomeres was usually not observed in these strains. Strains with the tel1 yku80, mec1 tel1 yku70, and mec1 tel1 yku80 genotypes, however, often yielded PCR fragments ≈500 bp in size (Fig. 3a). We cloned and sequenced four such fragments derived from mec1 tel1 yku80 strains. Two of the fusions involved poly G1–3 tracts derived from both X and Y′ telomeres, and two had poly G1–3 tracts derived from one of the subtelomeric repeats with ambiguous sequences at the other repeat. We conclude, therefore, the T-TFs occur in strains with the yku mutations, but the types of fusions are different from those observed in strains with wild-type Ku proteins. We also examined the rate of CAN1 deletions in strains with the yku mutations (Table 2). The triple mutants had deletion rates that were ≈10-fold lower than observed in the mec1 tel1 strain.
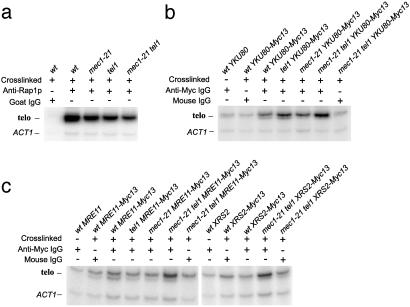
Interaction of Yku80p, Mre11p, and Xrs2p with Telomeric DNA in mec1 tel1 Strains. One explanation for T-TF formation in the mec1 tel1 strains is that the telomeres in this genetic background are devoid of telomere-binding proteins that protect the ends from fusions. We used chromatin immunoprecipitation techniques to examine telomere binding of epitope-tagged (13 × Myc epitope) Yku80p, Mre11p, and Xrs2p; MEC1 TEL1 strains with the tagged proteins had wild-type telomere length, indicating they had wild-type function. The telomere-binding Rap1p was used as a control.
Chromatin was isolated from exponentially growing cells. After immunoprecipitation of the crosslinked chromatin, the crosslinks were reversed, and the DNA was amplified by primers specific for the telomeres (resulting in a 258-bp fragment derived from the X element of chromosome XV) and a nontelomeric control fragment. As expected (23), anti-Rap1p antibody efficiently precipitated telomeric chromatin in wild-type, mec1, tel1, and mec1 tel1 strains (Fig. 4a). The reduction in intensity of the telomeric fragment in the tel1 and mec1 tel1 strains is expected because of smaller amount of double-stranded telomeric repeats (the binding site of Rap1p). Yku80p has been previously shown to bind yeast telomeres in vivo (24). The anti-Myc antibody precipitated telomeric sequences from Yku80-Myc-containing chromatin extracts derived from wild-type, mec1, tel1, and mec1 tel1 cells (Fig. 4b). The slightly higher level of the telomere-specific PCR fragment in the mec1 tel1 strain was reproducibly observed. Thus, the absence of the Tel1 and Mec1 activities does not reduce the binding of Yku80p.
Fig. 4.
Measurement of binding of Rap1p, Ku80p, Mre11p, and Xrs2p to telomeric sequences in wild-type, tel1, mec1–21, and mec1–21 tel1 strains by chromatin immunoprecipitation. Immunoprecipitated chromatin samples were analyzed by PCR with two pairs of primers: one pair that amplifies the DNA from the X repeat of chromosome XV (labeled “telo”) and one that amplifies a portion of the nontelomeric ACT1 gene. (a) Rap1p binding to telomeres. Strain names in lanes 1–5 are: JMY309-1a (lanes 1 and 2), JMY309-1b, JMY309-1c, and JMY309-1d. (b) Yku80p binding to telomeres. Strain names in lanes 1–7 are: JMY309-1a, PM116-6a (lanes 2 and 3), PM116-1a, PM116-1b, and PM116-3c (lanes 6 and 7). (c) Mre11p and Xrs2p binding to telomeres. Strain names in lanes 1–12 are: JMY309-1a (lanes 1 and 8), PM121-1b (lanes 2 and 3), PM121-9c, PM121-1d, PM121-10a (lanes 6 and 7), PM115-1b (lanes 9 and 10), and PM115-9a (lanes 11 and 12).
By using an anti-Myc antibody in extracts derived from strains with Mre11-Myc and Xrs2-Myc, we did not find significant enrichment for the telomeric PCR fragment relative to the control DNA fragment in strains with the wild-type, mec1,or tel1 genotypes (Fig. 4c). In addition, we observed approximately the same signal by using the antibody directed against mouse IgG. In strains of the mec1 tel1 genotype and either Mre11-Myc or Xrs2-Myc; however, we found an enrichment for telomeric sequences. The absence of the MRX complex from the telomeres in wild-type, mec1, and tel1 strains was surprising, because the equivalent complex is present at the telomeres in mammalian cells (25). It is possible that the complex is present at the yeast telomeres at a restrictive time during the cell cycle, as is observed in mammalian cells (25). Alternatively, the MRX complex may be excluded from the telomeres in most strains, binding only to DNA ends that are recognized as double-stranded DNA breaks (for example, the short telomeres characteristic of the mec1 tel1 genotype). Because the MRX complex binds telomeres in the mec1 tel1 strains, binding does not require Tel1p- or Mec1p-dependent phosphorylation of the MRX proteins; similarly, DNA damage-induced formation of the comparable complex in mammalian cells is independent of ATM (26).
Discussion
End-to-end chromosome associations are common in mammalian cells that have defects in telomere length regulation (1). Mutants affecting telomere metabolism result in chromosome circularization in Schizosaccharomyces pombe (27) and Kluyveromyces lactis (28). In S. cerevisiae, fusions between subtelomeric, but not telomeric (poly G1–3T/C1–3A), repeats are observed in an est2 strain (8), as are circular yeast chromosomes, presumably representing fusions between telomeric or subtelomeric repeats (9). In addition, Chan and Blackburn (10) found fusions between telomeric repeats and an HO-induced DNA break in tel1 est2 and tlc1 tel1 strains. Fusions between two telomeric repeats have not been described previously in S. cerevisiae, although such fusions have been seen in S. pombe (29) and in mammalian cells lacking TRF2 (30).
Because T-TFs have no sequence homology at the fusion breakpoint, they represent NHEJ events. We found that T-TF formation was independent of the Ku proteins and the MRX complex (proteins required for “normal” NHEJ) but required DNA ligase IV. In S. cerevisiae, fusions between an HO-induced break and telomeric sequences in tel1 and tel1 tlc1 strains are substantially reduced by lig4 mutations (10), and circular chromosome formation in est2 strains is Lig4p-dependent (9). In addition, in mammalian cells with nonfunctional TRF2, telomere fusion events are ligase IV-dependent (30). In S. pombe, however, the lig4+ gene product is not necessary for chromosome circularization (fusing two subtelomeric repeats) associated with deletion of trt1+ (31).
Addition of the yku70 or yku80 mutations to the mec1 tel1 strain greatly reduced the frequency of 350 bp T-TF-specific PCR product but resulted in increased levels of a larger T-TF fragment. It is possible that the formation of the 350 bp T-TF requires blunt DNA ends, a substrate that is not present in strains with the yku mutations (32). The larger T-TF product observed in the mec1 tel1 yku70 or mec1 tel1 yku80 strains may reflect a small fraction of telomeres involved in a telomerase-independent elongation process that generates blunt ends. Because mutations in the KU genes result in telomere-telomere associations in mammalian cells (33) and plants (34), the Ku independence of T-TFs appears general.
Correlation Between the T-TFs and CAN1 Deletions. There is good agreement between the frequency of T-TFs and the level of genome instability as measured by the rate of CAN1 deletions. The highest rates of CAN1 deletions (>3 × 10–6 per cell division) were observed in mec1 tel1, mec3 tel1, and ddc1 tel1 strains, which also had the highest levels of T-TFs. The lowest rates of CAN1 deletions (<10–7 per cell division) were found in strains with relatively low rates of T-TFs. In general, the relative frequencies of T-TFs were similar when estimated by the semiquantitative PCR procedure and real-time PCR.
Our results are also in good agreement with other assays of genome instability (codeletion of two linked markers) done by Kolodner and coworkers (6, 17, 35). High rates (>1,000× wild type) of codeletions were observed in mec1 tel1, mec3 tel1, ddc1 tel1, and pds1 tel1 strains, genotypes for which we detected T-TFs. Lower deletion rates (200-fold or less) were observed for mec1 sml1, tel1, mec1 tlc1, tlc1, and mec1 yku70 strains, which had low levels of T-TFs. The only substantive difference between our data and these previous studies is that we found that pds1 tel1 had lower levels of T-TFs and lower rates of deletions than the mec1 tel1 strain.
The correlation between T-TF levels and genome stability in various mutant strains and the finding that genetic alterations that reduce the frequency of T-TFs in mec1 tel1 strains (expression of the Cdc13-Est2 fusion or addition of the lig4 mutation) also reduce the frequency of CAN1 deletions, suggest that T-TFs are an intermediate in the production of the deletions. T-TFs will result in dicentric chromosomes (if the fusions involve different chromosomes or sister chromatids) or circular chromosomes (if the fusions involve a single chromosome). Both types would be expected to result in subsequent double-strand breaks, and NHEJ events involving these broken chromosomes would generate chromosome rearrangements characteristic of the mec1 tel1 genotype. Similar models have been proposed by other researchers to account for related observations.
Mechanism of T-TF Formation. The highest levels of T-TFs were found in strains with: (i) short “uncapped” telomeres, (ii) wild-type DNA ligase IV, and (iii) a checkpoint deficiency. Telomeres have a “cap” that protects them from engaging in DNA repair events such as fusions (1). The composition of this cap in S. cerevisiae is unclear, but Tel1p is involved directly or indirectly in the capping function (10, 36). Because we find that mec1 tel1 strains have increased binding of MRX proteins to the telomeres, the Tel1p/Mec1p activities are not required to promote a cap function of the MRX complex. In the absence of the capping function, the telomeres in mec1 tel1 strains and related genotypes are likely to be susceptible to ligation by DNA ligase IV. We assume that the ligation event occurs between two DNA telomeres with blunt ends. Because T-TFs occur between long telomeres in the mec1 tel1 yku strains, defective cap formation is more important for T-TF formation than short telomeres per se.
The highest level of T-TFs are observed in strains with a mutation in TEL1 and a mutation in one of the MEC1, DDC1, or MEC3 DNA damage checkpoint genes. Mec1p is involved in almost every DNA damage checkpoint, whereas Ddc1 and Mec3p form a proliferating cell nuclear antigen-like clamp specialized for DNA repair synthesis (20, 37). Although we cannot rule out a direct role of the checkpoint genes in telomere elongation or capping, it is likely that their role in T-TF formation is a consequence of their checkpoint functions. Enomoto et al. (38) argued the existence of a checkpoint that monitored telomere length and depended on the Mec3, Mec1, and Ddc1 proteins. A second possibility is that short telomeres are detected as DNA damage by the checkpoint pathway in the same manner as double-stranded DNA breaks. The last possibility is that the checkpoint monitors the existence of T-TFs indirectly, by detecting the dicentric chromosome, DNA breaks resulting from processing the dicentric chromosome, or the existence of the palindromic structure resulting from the T-TF.
In conclusion, strains with short telomeres and a DNA damage checkpoint deficiency have high levels of T-TFs and high rates of deletions. Because similar types of instability are seen in mammalian cells with short telomeres, S. cerevisiae is a useful organism for investigating the genomic chaos resulting from T-TFs.
Supplementary Material
Acknowledgments
We thank R. Craven, J. Mallory, K. Ritchie, T. de Lange, E. Louis, and D. Gottschling for discussions; and R. Rothstein (Columbia University, New York), S. Elledge (Harvard University, Cambridge, MA), Y. Sanchez (University of Cincinnati College of Medicine, Cincinnati), V. Lundblad (Baylor College of Medicine, Houston), R. Kolodner (Ludwig Institute for Cancer Research, La Jolla, CA), D. Gottschling (Fred Hutchinson Cancer Research Center, Seattle), R. Wellinger (Université de Sherbrooke, Sherbrooke, PQ, Canda), P. Greenwell (University of North Carolina, Chapel Hill), M. Barbara (University of North Carolina, Chapel Hill), and R. Craven (University of Kentucky, Lexington) for strains. The research was supported by National Institutes of Health Grant GM52319.
Abbreviations: T-TF, telomere-telomere fusion; NHEJ, nonhomologous end-joining.
References
- 1.de Lange, T. (2002) Oncogene 21, 532–540. [DOI] [PubMed] [Google Scholar]
- 2.Pandita, T. K. (2002) Oncogene 21, 611–618. [DOI] [PubMed] [Google Scholar]
- 3.Hackett, J. A. & Greider, C. W. (2002) Oncogene 21, 619–626. [DOI] [PubMed] [Google Scholar]
- 4.Ritchie, K. B., Mallory, J. C. & Petes, T. D. (1999) Mol. Cell. Biol. 19, 6054–6075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chan, S. W. L., Chang, J., Prescott, J. & Blackburn, E. H. (2001) Curr. Biol. 11, 1240–1250. [DOI] [PubMed] [Google Scholar]
- 6.Myung, K., Datta, A. & Kolodner, R. D. (2001) Cell 104, 397–408. [DOI] [PubMed] [Google Scholar]
- 7.Craven, R. J., Greenwell, P. W., Dominska, M. & Petes, T. D. (2002) Genetics 161, 493–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hackett, J. A., Feldser, D. M. & Greider, C. W. (2001) Cell 106, 275–286. [DOI] [PubMed] [Google Scholar]
- 9.Liti, G. & Louis, E. J. (2003) Mol. Cell 11, 1373–1378. [DOI] [PubMed] [Google Scholar]
- 10.Chan, S. W.-L. & Blackburn, E. H. (2003) Mol. Cell 11, 1372–1387. [Google Scholar]
- 11.Thomas, B. J. & Rothstein, R. (1989) Genetics 123, 725–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wierdl, M., Greene, C. N., Datta, A., Jinks-Robertson, S. & Petes, T. D. (1996) Genetics 143, 713–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sanchez, Y., Desany, B., Jones, W. J., Liu, Q., Wang, B. & Elledge, S. J. (1996) Science 271, 357–360. [DOI] [PubMed] [Google Scholar]
- 14.Kironmai, K. M. & Muniyappa, K. (1997) Genes Cells 2, 443–455. [DOI] [PubMed] [Google Scholar]
- 15.Boulton, S. J. & Jackson, S. P. (1998) EMBO J. 17, 1819–1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ritchie, K. B. & Petes, T. D. (2000) Genetics 155, 475–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Myung, K., Chen, C. & Kolodner, R. D. (2001) Nature 411, 1073–1076. [DOI] [PubMed] [Google Scholar]
- 18.Tsukamoto, Y., Taggart, A. K. P. & Zakian, V. A. (2001) Curr. Biol. 11, 1328–1335. [DOI] [PubMed] [Google Scholar]
- 19.Evans, S. K. & Lundblad, V. (1999) Science 286, 117–120. [DOI] [PubMed] [Google Scholar]
- 20.Nyberg, K. A., Michelson, R. J., Putnam, C. W. & Weinert, T. A. (2002) Annu. Rev. Genet. 36, 617–656. [DOI] [PubMed] [Google Scholar]
- 21.Critchlow, S. E. & Jackson, S. P. (1998) Trends Biochem. Sci. 23, 394–398. [DOI] [PubMed] [Google Scholar]
- 22.Valencia, M., Bentele, M., Vaze, M. B., Herrmann, G., Kraus, E., Lee, S. E., Schar, P. & Haber, J. E. (2001) Nature 414, 666–669. [DOI] [PubMed] [Google Scholar]
- 23.Conrad, M. N., Wright, J. H., Wolf, A. J. & Zakian, V. A. (1990) Cell 16, 739–750. [DOI] [PubMed] [Google Scholar]
- 24.Martin, S. G., Laroche, T., Suka, N., Grunstein, M. & Gasser, S. M. (1999) Cell 97, 621–633. [DOI] [PubMed] [Google Scholar]
- 25.Zhu, X.-D., Kuster, B., Mann, M., Petrini, J. H. J. & de Lange, T. (2000) Nat. Genet. 25, 347–352. [DOI] [PubMed] [Google Scholar]
- 26.Mirzoeva, O. K. & Petrini, J. H. J. (2001) Mol. Cell. Biol. 21, 281–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nakamura, T. M., Moser, B. A. & Russell, P. (2002) Genetics 161, 1437–1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McEachern, M. J., Iyer, S., Fulton, T. B. & Blackburn, E. H. (2000) Proc. Natl. Acad. Sci. USA 97, 11409–11414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ferreira, M. & Cooper, J. (2001) Mol. Cell 7, 55–63. [DOI] [PubMed] [Google Scholar]
- 30.Smogorzewska, A., Karlseder, J., Holtgreve-Grez, H., Jauch, A. & de Lange, T. (2002) Curr. Biol. 12, 1635–1644. [DOI] [PubMed] [Google Scholar]
- 31.Baumann, P. & Cech, T. R. (2000) Mol. Biol. Cell 11, 3265–3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gravel, S., Larrivee, M., Labrecque, P. & Wellinger, R. J. (1998) Science 280, 741–744. [DOI] [PubMed] [Google Scholar]
- 33.Goytisolo, F. A. & Blasco, M. A. (2002) Oncogene 21, 584–591. [DOI] [PubMed] [Google Scholar]
- 34.Riha, K. & Shippen, D. E. (2003) Proc. Natl. Acad. Sci. USA 100, 611–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Myung, K. & Kolodner, R. D. (2002) Proc. Natl. Acad. Sci. USA 99, 4500–4507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.DuBois, M. L., Haimberger, Z. W., McIntosh, M. W. & Gottschling, D. E. (2002) Genetics 161, 995–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Majka, J. & Burgers, P. M. J. (2003) Proc. Natl. Acad. Sci. USA 100, 2249–2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Enomoto, S., Glowczewski, L. & Berman, J. (2002) Mol. Biol. Cell 13, 2626–2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.