Abstract
CD1d-restricted natural killer T (NKT) cells are a subset of regulatory T cells that react with glycolipid antigens. Although preclinical studies have effectively targeted NKT cells for immunotherapy, little is known regarding the early in vivo response of these cells to antigenic stimulation. We have analyzed the early response of NKT cells to glycolipid antigens and bacterial infection by using specific reagents for tracking these cells. Our results demonstrate dramatic in vivo expansion and surface phenotype alterations after NKT cell activation with α-galactosylceramide. In addition, we show significant NK1.1 down-modulation on NKT cells in the setting of oral Salmonella infection. Our results indicate that in vivo activation of NKT cells leads to a dynamic response characterized by surface receptor down-modulation and expansion. These findings alter current understanding of NKT cell biology and should aid in the rational design of NKT cell-based immunotherapies.
Natural killer T (NKT) cells are a group of T lymphocytes with surface markers and characteristics typical of innate immune cells (1, 2). Prototypical NKT cells express an invariant Vα14-Jα18/Vβ8 (Vα24-Jα18/Vβ11 in human) T cell receptor (TCR), together with the NK cell marker NK1.1. NKT cells are specific for glycolipid antigens such as the marine sponge-derived agent α-galactosylceramide (α-GalCer) presented by the major histocompatibility complex class I-like molecule CD1d (1, 2). NKT cells have been implicated in a variety of immune responses, playing largely a regulatory role (1, 2). Because of their immunomodulatory properties, NKT cells are attractive targets for the development of immunotherapies (3).
Despite promising preclinical studies with NKT cell agonists such as α-GalCer, little is known regarding the early in vivo effects of NKT cell activation on the cellular proliferation of NKT cells themselves. A number of studies have shown that quickly (within 8–12 h) after their activation, NKT cells become undetectable (4–12). The apparent demise of NKT cells has been observed in the context of in vivo anti-CD3 (4), α-GalCer (5–8), and IL-12 (4) stimulation, and in the setting of viral (9, 10) and bacterial (11, 12) infection. This phenomenon has been correlated with increased apoptosis and increased expression of Fas and FasL by NKT cells (4, 13), suggesting that NKT cells are exquisitely sensitive to Fas-mediated activation-induced cell death (AICD). According to current models of NKT cell activation, the demise of NKT cells in the liver and spleen is quickly (within 48 h) followed by repopulation of these cells due to homeostatic proliferation of an NKT cell reservoir in the bone marrow (4). However, this model is at variance with a number of other studies on NKT cells, such as their capacity to proliferate robustly in response to TCR engagement in vitro (14, 15) and the capacity of α-GalCer to modulate the quality of NKT cell responses to subsequent exposures to this antigen (3, 7, 16–18). The current models of NKT cell activation also raise mechanistic questions regarding the benefits of multiple α-GalCer dosings that have been used in preclinical therapeutic protocols (3).
Here we have reevaluated the in vivo population dynamics of NKT cells in response to glycolipid antigen stimulation and bacterial infection by using specific assays for identifying and quantifying these cells ex vivo. Our findings warrant a revision of current models of NKT cell activation, will guide future investigations regarding NKT cell responses, and should aid in the design of NKT cell-based therapeutic protocols.
Materials and Methods
Mice. Eight- to 10-week-old female C57BL/6J (B6) mice, thymectomized and sham-thymectomized B6 mice, and B6.TCRα–/– mice were purchased from The Jackson Laboratory. B6.Vα14 transgenic mice (19) were provided by A. Bendelac (University of Chicago, Chicago).
Reagents. A synthetic form of α-GalCer (KRN7000) (20) was obtained from the Kirin Brewery (Gunma, Japan). For most experiments, mice were treated with 5 μg of α-GalCer by i.p. injection in a final volume of 200 μl of PBS. Anti-CD3, anti-CD3-PerCPCy5.5, anti-TCRβ–allophycocyanin (APC) and -FITC, anti-NK1.1-APC and -phycoerythrin, and anti-B220-PerCP antibodies were purchased from PharMingen. Tetrameric CD1d molecules loaded with α-GalCer were prepared as described (6, 21). Carboxyfluorescein diacetate succinimidyl ester (CFSE) was obtained from Molecular Probes.
Flow Cytometry. Spleen and liver mononuclear cells were stained as described (18). In some experiments, B220+ and dead cells (propidium iodide-positive) were excluded from the analysis by electronic gating. Intracellular staining was performed with Cytofix/Cytoperm reagents and Golgi-stop, purchased from PharMingen, according to the manufacturer's specifications. Data acquisition was performed on a FACSCalibur instrument with cellquest software (Becton Dickinson), and data were analyzed by using flowjo software. The percentage of surface TCR expression in TCR down-modulation experiments was calculated as follows: 100 × (geometric MFI of tetramer staining among gated NKT cells in the experimental condition-geometric MFI of tetramer staining among the nonstaining population)/(geometric MFI of tetramer staining among gated NKT cells in the control condition–geometric MFI of tetramer staining among the nonstaining population) (MFI, mean fluorescence intensity).
Quantitative Real-Time PCR Analysis. PCRs were performed on an ABI Prism 7700 real-time PCR machine (Applied Biosystems). SYBR green PCR Master Mix and TaqMan Reverse Transcriptase reagents were purchased from Applied Biosystems and used according to the manufacturer's specifications. Primers for the amplification of Vα14-Jα18 TCR rearrangements and cDNA corresponding to reverse-transcribed IL-4, IL-10, and IFN-γ have been described (22, 23). Analysis of real-time PCR data was performed according to the manufacturer's instructions. β-Actin was used as the internal standard. β-Actin primers were as described (23).
Proliferation and Cytokine Production Assays. Splenocytes (2 × 105) were incubated with titrated doses of α-GalCer in RPMI 1640 medium supplemented with 10% FCS/50 μM 2-mercaptoethanol/2 mM glutamine/antibiotics/10 mM Hepes (complete medium) for 72 h. For proliferation assays, 1 μCi of [3H]thymidine (New England Nuclear) was then added to the wells, and after an additional 16 h of culture, cells were harvested with a cell harvester (Tomtec, Orange, CT), and uptake of radioactivity was measured with a betaplate reader (Wallac, Gaithersburg, MD). For measurement of cytokine levels, culture supernatants were collected, and cytokine levels were determined by ELISA, as described (18).
In Vitro TCR Down-Modulation and CFSE Dilution Assays. For in vitro TCR down-modulation experiments, cells were plated in complete medium and, at the appropriate time points, α-GalCer was added to a final concentration of 100 ng/ml. Cells were then briefly centrifuged (2 min, 500 × g) to facilitate rapid NKT cell contact with antigen-presenting cells. Anti-CD3 antibodies were coated onto round-bottom 96-well plates for 90 min at 37°C at a concentration of 10 μg/ml in 30 μl of PBS. Wells were washed three times with cold PBS, cells were added, and plates were centrifuged as above. For in vitro expansion experiments (Figs. 2 and 3F), cells were cultured for 24 h with or without 100 ng/ml α-GalCer. After 24 h, α-GalCer-containing medium was replaced with fresh culture medium without α-GalCer, and the cultures were continued as indicated in Fig. 2 and 3 legends until flow cytometric analysis. For CFSE dilution experiments, the Vybrant CFDA SE cell Tracer Kit was purchased from Molecular Probes and used according to the manufacturer's specifications.
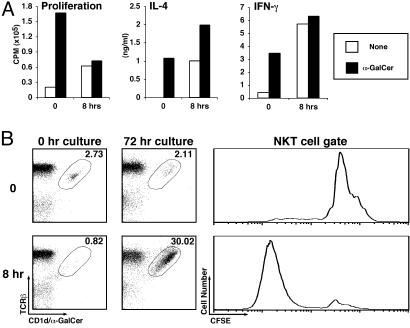
Fig. 2.
Reappearance of NKT cells on in vitro culture. (A) Spleen cells from uninjected mice (0) and from mice injected 8 h earlier with α-GalCer (5 μg, i.p.) were cultured in vitro in the presence or absence of α-GalCer (50 ng/ml). Three days later, proliferation was measured by [3H]thymidine uptake, and IL-4 and IFN-γ levels were measured by ELISA. (B) Splenocytes were harvested at the indicated time points after α-GalCer administration (5 μg, i.p.), labeled with CFSE, and cultured in vitro without further stimulation for 72 h. NKT cells were analyzed by flow cytometry before and after culture. Numbers indicate the percentage of TCRβ+ CD1d/α-GalCer tetramer+ cells among B220– cells. CFSE plots on tetramer+ cells were generated for cells cultured for 72 h.
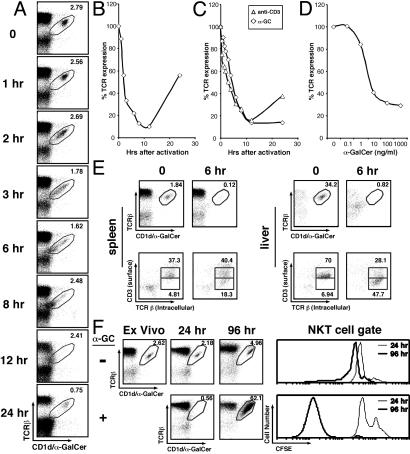
Fig. 3.
NKT cells quickly down-regulate their surface TCRs on activation. (A and B) Spleen cells from mice injected i.p. with 5 μgof α-GalCer were killed at the indicated time points and stained with anti-TCRβ antibodies and CD1d/α-GalCer tetramers (A). The percentage of TCR expression for cells gated in A is expressed relative to tetramer binding on cells from uninjected mice (B). (C) Kinetic analysis of TCR surface down-regulation on NKT cells activated in vitro with 100 ng/ml of α-GalCer or plate-bound anti-CD3 antibodies. (D) Analysis of TCR surface down-regulation on NKT cells activated in vitro for 6 h with titrated doses of α-GalCer. TCR surface levels are expressed as the percentage of TCR surface expression on unstimulated NKT cells. (E) Spleen and liver mononuclear cells were harvested from uninjected mice and 6 h after i.p. injection of 5 μgof α-GalCer, and cells were stained with anti-CD3-PerCP-Cy5.5 and anti-NK1.1-allophycocyanin antibodies. An excess of unlabeled anti-TCRβ antibodies were added to the staining mixture to saturate surface TCRs. Cells were then fixed and permeabilized, stained with anti-TCRβ–phycoerythrin antibodies, and analyzed by flow cytometry. Flow cytometry plots represent cells electronically gated for intermediate levels of NK1.1 expression (Lower), which is typical of NKT cells (e.g., see Fig. 1 A and C). TCRβint CD1d/α-GalCer tetramer+ cell populations were identified in parallel stainings (Upper). (F) Recapitulation of the dynamics of NKT cell activation in vitro. Spleen cells were labeled with CFSE, cultured without (–) or with (+) 100 ng/ml α-GalCer for 24 h, then washed and cultured for an additional 72 h. Cells were stained at the 0-, 24-, and 96-h time points with anti-TCRβ and anti-B220 antibodies and CD1d/α-GalCer tetramers and analyzed by flow cytometry as in Fig. 1B. CFSE plots are shown for TCRβint CD1d/α-GalCer tetramer+ cells from the 24- (thin lines) and 96- (heavy lines) h time points.
Enrichment of NK1.1+ and NK1.1– NKT Cells. Spleen cells were harvested from Vα14 transgenic mice, stained with anti-NK1.1-phycoerythrin (PE) antibodies, and NK1.1+ cells were positively selected by magnetic-activated cell separation by using anti-PE magnetic beads (Miltenyi Biotec). CD4+ cells were positively selected from the flow-through by using anti-CD4 magnetic beads.
Salmonella Infections. Doses of 1–2 × 109 bacteria (Salmonella typhimurium χ4666/OVA SD) were inoculated by the oral route in 9- to 12-week-old B6 mice, as described (12). Spleens were harvested at different days postinfection. Bacterial load was determined by plating serial dilutions of single-cell suspensions on Luria–Bertoni agar plates (12).
Results
NKT Cell Population Dynamics After in Vivo Activation with α-GalCer. We focused on the response of NKT cells to α-GalCer, because this reagent has great potential as an immunotherapeutic (3). For most of our studies, we selected a dose (5 μg) and injection route (i.p.) commonly used in α-GalCer-based treatment protocols in mice. For identification of invariant NKT cells, we used anti-TCRβ and -NK1.1 antibodies, as well as tetrameric CD1d molecules loaded with α-GalCer (CD1d/α-GalCer tetramers) (6, 21, 24).
Consistent with prior studies (5), analysis with anti-TCRβ and -NK1.1 antibodies demonstrated loss of TCRβint NK1.1+ cells from the spleen within8hof α-GalCer injection (Fig. 1A). These cells reappeared 48 h after injection, returning to normal or slightly enhanced levels after 7 days. Similar staining patterns were obtained in other organs, including liver, peripheral blood, and bone marrow (see Figs. 6–8, which are published as supporting information on the PNAS web site, www.pnas.org). In agreement with prior studies (5), cells with a TCRβ– NK1.1+ surface phenotype typical of classical NK cells were also transiently depleted from the spleen (Fig. 1 A and B) ≈24 h after α-GalCer administration.
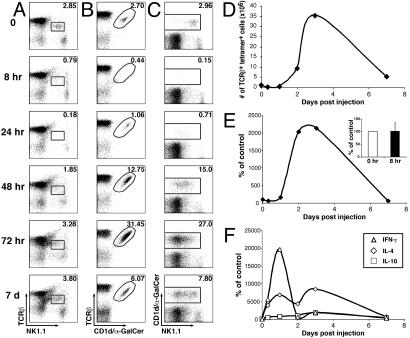
Fig. 1.
NKT cell population dynamics in the spleen after in vivo activation with α-GalCer. (A–D) Splenic NKT cell populations were analyzed by flow cytometry at the indicated times after i.p. injection of 5 μg of α-GalCer. Numbers in A–C indicate the average percentage of TCRβint NK1.1+ (A), TCRβint CD1d/α-GalCer tetramer+ (B), and NK1.1+ CD1d/α-GalCer tetramer+ (C) cells among B220– lymphocytes of two mice at each time point. The average absolute numbers of TCRβint CD1d/α-GalCer tetramer+ cells in the spleen of two α-GalCer-injected mice at each time point were calculated (D). (E) DNA was harvested from spleen tissue at different time points after in vivo activation and Vα14-Jα18 DNA copy number was measured by quantitative real-time PCR. (Inset) Real-time PCR data averaged for three independent experiments at the 0- and 8-h time points. (F) Total RNA was harvested from spleen tissue at different time points after injection of α-GalCer and analyzed for IFN-γ, IL-4, and IL-10 mRNA expression levels by quantitative real-time RT-PCR.
Results for staining with anti-TCRβ antibodies and CD1d/α-GalCer tetramers to identify NKT cells showed a dramatically different picture (Fig. 1 B and D). Although TCRβint tetramer+ cells quickly disappeared from the spleen after α-GalCer injection, this population began to reappear 24 h after injection and was dramatically increased by 48 h (≈5-fold) and 72 h (>10-fold); the prevalence of NKT cells remained slightly enhanced 7 days after injection. Robust NKT cell expansion (5- to 15-fold) was observed at high doses of α-GalCer (0.5–10 μg per mouse), whereas expansion was less profound at lower doses of this reagent (data not shown). Similar population dynamics were observed in the liver, bone marrow, and peripheral blood, but expansion was less profound in the liver and bone marrow as compared with spleen and peripheral blood (compare Figs. 1B, 6B, 7B, and 8B). Thus, the apparent population dynamics of NKT cells depend on the specific reagents used to detect these cells.
Robust in vivo expansion of NKT cell populations in response to TCR engagement has not been previously documented. Because expansion was observed only with CD1d/α-GalCer tetramers, we wanted to confirm this finding by methods independent of cell surface markers. Therefore, we measured the relative numbers of NKT cells by using a quantitative real-time PCR assay to detect the presence of rearranged Vα14-Jα18 TCRα chains among splenic DNA. Our results indicated profound (≈20-fold) NKT cell expansion at days 2 and 3 (Fig. 1E).
Persistence of NKT Cells After in Vivo Activation. In addition to providing information on NKT cell expansion, the real-time PCR assay permitted us to reevaluate the fate of NKT cells early after in vivo TCR engagement. We found that, at the 8-h time point after α-GalCer injection, when NKT cells are undetectable by anti-NK1.1 antibodies and tetramers (Fig. 1 A–D), numbers of genomic Vα14-Jα18 rearrangements are indistinguishable from those in spleens of uninjected mice (see Fig. 1E Inset). Thus, these findings suggest that a significant fraction of NKT cells resists AICD and persists in the spleen.
To provide additional evidence for the conclusion that NKT cells persist in the spleen of α-GalCer-injected mice, we evaluated the kinetics of cytokine mRNA expression at different time points after α-GalCer injection. Results showed expression of multiple cytokine transcripts, including IFN-γ, IL-4, and IL-10, for several days (Fig. 1F). Interestingly, IL-4 gene expression was also detected at early time points (8 h) when TCRβint NK1.1+ and tetramer+ cells are not detected. Because NKT cells are the main, if not exclusive, source of IL-4 early in the response to α-GalCer stimulation (14), these findings further confirm the presence of NKT cells in the spleen 8 h after α-GalCer administration.
If NKT cells indeed persist in the spleen 8 h after α-GalCer injection, they would be expected to proliferate and produce cytokines on in vitro culture. To test this possibility, we harvested spleen cells from uninjected mice and from mice injected with α-GalCer 8 h earlier. Cells were then cultured in the absence or presence of α-GalCer and evaluated for proliferation and IFN-γ and IL-4 production. As expected, cells from uninjected mice proliferated and produced cytokines only in the presence of α-GalCer (Fig. 2 A). In sharp contrast, we observed robust proliferation and cytokine production in spleen cultures prepared 8 h after α-GalCer injection, even in the absence of additional antigenic stimulation. Interestingly, IL-4 production in the latter cultures was enhanced on addition of α-GalCer. Although NK cells may be the main source of IFN-γ production in these cultures (5, 25), NKT cells themselves are the predominant source of IL-4 production (14). Hence, NKT cells, although undetectable by surface markers, are present in the spleens of mice 8 h after α-GalCer injection.
Because a variety of cell types may be proliferating in these cultures, we analyzed them by flow cytometry. Results demonstrated profound expansion of TCRβint tetramer+ cells in cultures from the 8-h time point after in vivo α-GalCer injection (Fig. 2B). CFSE dilution experiments further showed that the population of expanded NKT cells had divided extensively (Fig. 2B).
Prior studies have argued that NKT cells undergo massive cell death in the spleen and liver as quickly as 2 h after TCR engagement and that few, if any, NKT cells persist at 8–24 h (4). Our findings indicate that, whereas a significant fraction of NKT cells undergo AICD (data not shown), a pool of NKT cells persists in the spleen (Figs. 1 and 2) and liver (Fig. 6) and undergoes multiple rounds of rapid proliferation, without the need for a reservoir of NKT cells outside of these organs.
Activated NKT Cells Down-Regulate Surface TCR Expression. To gain insight into the mechanism by which antigen-stimulated NKT cells are rendered undetectable by CD1d/α-GalCer tetramers, we considered the possibility that NKT cells undergo rapid TCR down-regulation, as described for conventional T cells stimulated with anti-CD3 antibodies or specific antigens (26). Thus, we analyzed surface TCR levels on NKT cells at early time points after α-GalCer injection. We observed a gradual decrease in the MFI of tetramer staining after α-GalCer administration (Fig. 3 A and B). TCR down-modulation was detected within 1 h and was maximal at 8–12 h, after which TCR expression levels increased again, returning to normal levels at 24–48 h (Figs. 1 A–C and 3B). Similar results were obtained in vitro by using NKT cells stimulated with α-GalCer or plate-bound anti-CD3 antibodies (Fig. 3C). These kinetics of TCR down-regulation on NKT cells are reminiscent of those observed for antigen-stimulated conventional T cells (27). In addition, dose–response studies showed that TCR down-regulation on NKT cells is enhanced by increasing amounts of α-GalCer in the culture medium (Fig. 3D).
For conventional T cells, TCR surface down-regulation results from decreased surface recycling of internalized TCR complexes (28). Therefore, we assayed extra- and intracellular pools of TCR expression in α-GalCer-activated NKT cells by flow cytometry. Results showed that 6 h after α-GalCer injection, a significant fraction of NKT cells in the spleen and especially in the liver contain intracellular but little surface TCR (Fig. 3E). We conclude that the NKT cell population that escapes tetramer staining in the spleen and liver at early time points after α-GalCer injection can be visualized by intracellular staining for TCR expression.
In Vitro Recapitulation of the Surface Marker Down-Regulation/Expansion Model of NKT Cell Activation. Thus far, our results indicate that antigen-stimulated NKT cells quickly down-regulate their TCR, rapidly proliferate, resume surface TCR expression, and become apparent in large numbers by tetramer but not anti-NK1.1 staining at 48–72 h. To provide further support for this surface marker down-regulation/expansion model of NKT cell activation, we attempted to recapitulate it in a completely in vitro setting. Our results showed that, in α-GalCer-stimulated cultures, NKT cells are undetectable by tetramer staining 24 h after culture but are dramatically expanded at the 96-h time point (Fig. 3F). In addition, expanded NKT cells undergo NK1.1 down-regulation (e.g., see Fig. 4C Inset). CFSE dilution experiments showed that NKT cells had undergone numerous rounds of cell division (Fig. 3F). These findings indicate that the in vivo behavior of NKT cells to α-GalCer stimulation can be closely mimicked in vitro.
Fig. 4.
Precursors for NKT cell expansion. (A) Thymectomized and shamthymectomized mice were injected 1 mo postsurgery with α-GalCer (5 μg, i.p.), killed at the indicated time points, stained, and analyzed by flow cytometry as in Fig. 1B.(B) NK1.1+ and CD4+NK1.1– cells were isolated from spleens of Vα14 transgenic mice by magnetic beads as described in Materials and Methods. These two populations were placed in parallel cultures with spleen antigen-presenting cells from TCRα–/– mice in the presence of 100 ng/ml of α-GalCer. Cells were analyzed for down-modulation of surface TCRs as in Fig. 3A, and data were plotted as in Fig. 3B. (C) Enriched NK1.1+ and CD4+NK1.1– cells were labeled with CFSE and cultured as in B. After 24 h, α-GalCer-containing medium was replaced with fresh medium lacking α-GalCer for the remainder of the culture period. CFSE dilution among gated CD1d/α-GalCer tetramer+ NKT cells was assessed in parallel cultures of isolated NK1.1– and NK1.1+ cells at the indicated time points. Dot plots within the histograms show levels of NK1.1 expression (y axis) vs. CFSE dilution (x axis) in cultures of the NK1.1+ fraction of cells.
Precursor Populations for NKT Cell Expansion. We considered the possibility that preferential proliferation of an NK1.1-negative precursor population results in the generation of an expanded pool of NK1.1– NKT cells. Recent studies have identified such an NK1.1– NKT cell population with high proliferative capacity, representing recent thymic emigrants (29, 30). We therefore tested the in vivo response of thymectomized mice to α-GalCer. Our results revealed that NKT cell expansion in thymectomized and sham-thymectomized animals is indistinguishable (Fig. 4A). We further showed that purified NK1.1+ and NK1.1– NKT cell populations show similar TCR down-modulation kinetics (Fig. 4B) and proliferation potential (Fig. 4C) in response to in vitro α-GalCer stimulation, independent of extrasplenic NKT cell reservoirs. These results indicate that NKT cell expansion after antigen stimulation does not depend on NK1.1– precursors.
As an alternative explanation for the lack of NK1.1 expression on the expanded NKT cells, we evaluated the possibility that NKT cells down-regulate this marker for an extended time period after activation. Prior in vitro studies have shown that NKT cells lose NK1.1 expression on activation (31), but this has not been established in vivo. Analysis of NKT cells with anti-NK1.1 antibodies and CD1d/α-GalCer tetramers indicated that NKT cells progressively down-regulate the NK1.1 marker (Figs. 1C, 6C, 7C, and 8C). Thus, sustained down-regulation of the NK1.1 marker on activated NKT cells provides an explanation for the failure of prior studies to detect significant NKT cell expansion in vivo.
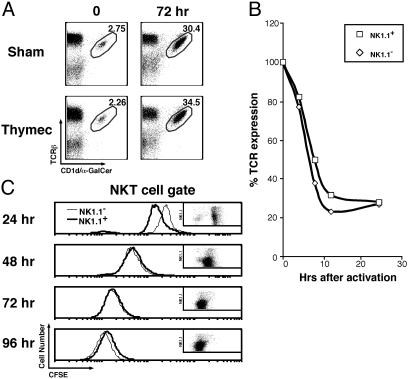
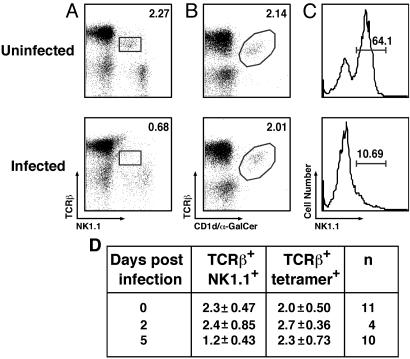
Dynamics of NKT Cells in the Setting of Bacterial Infection. Extensive down-modulation of surface markers on NKT cells may provide an explanation for the apparent loss of NKT cells after infection by certain microorganisms (9–12). Therefore, we reevaluated this issue using mice orally infected with S. typhimurium. Consistent with prior studies (12), the number of TCRβint NK1.1+ cells in the spleen dropped quickly and became almost undetectable 5 days after infection (Fig. 5 A and D). However, CD1d/α-GalCer tetramers permitted detection of the NKT cell population at this time point (Fig. 5 B and D). Evaluation of NK1.1 expression on tetramer+ cells revealed profound down-regulation of the NK1.1 marker (Fig. 5C). Hence, the apparent loss of NKT cells observed early after oral Salmonella infection is due, at least in part, to the loss of NK1.1 marker expression.
Fig. 5.
NKT cell dynamics during oral Salmonella infection. Control mice (uninfected) and mice infected with S. typhimurium were killed at 0, 2, or 5 days postinfection, and spleen cells were stained and analyzed by flow cytometry as in Fig. 1 A–C. Representative data at 5 days postinfection are shown in A–C. Numbers indicate the percentage of TCRβint NK1.1+ (A) and TCRβint CD1d/α-GalCer tetramer+ (B) cells among B220– lymphocytes for one uninfected and the average of three infected mice. NK1.1 levels on TCRβint tetramer+ cells were also evaluated (C). The data from several separate experiments are summarized in D.
Discussion
We have demonstrated several surprising findings regarding the response of NKT cells to in vivo activation. First, quickly after activation with an i.p. dose of 5 μg α-GalCer, NKT cells down-regulate their surface TCR, resulting in the apparent “disappearance” of these cells as observed with reagents directed against cell surface markers. Second, NKT cells are capable of robust in vivo proliferation, resulting in a dramatically (>10-fold) expanded NKT cell population in the spleen 3 days after α-GalCer injection. Third, the failure of prior studies to detect significant in vivo expansion of NKT cells is because of the lack of NK1.1 expression on this expanded NKT cell population. Fourth, NKT cells down-regulate NK1.1 expression after oral Salmonella infection. Thus, our findings warrant a reevaluation of current models of NKT cell activation.
Our results indicate that down-regulation of TCR expression provides an explanation for the apparent loss of NKT cells after in vivo TCR engagement. TCR down-regulation after antigenic stimulation is a well established phenomenon for conventional T cells (26) and may be a means for protecting T cells against overstimulation and AICD (27). In keeping with these findings, TCR down-regulation by NKT cells may protect these cells against overstimulation and prevent extensive tissue damage. Uncontrolled NKT cell responses can cause liver damage (32, 33) and induce abortions in mice (34). TCR down-modulation may provide a mechanism to avoid these undesirable consequences of NKT cell activation and rescue these cells from AICD.
Prior studies on NKT cell activation have failed to detect significant in vivo expansion of these cells. We were able to detect NKT cell expansion by using CD1d/α-GalCer tetramers and by a real-time PCR assay that quantifies rearranged Vα14-Jα18 TCRα chains among DNA. However, this expanded NKT cell population expresses little surface NK1.1, explaining why expansion has escaped detection in prior studies. We considered two alternative possibilities for the lack of NK1.1 expression on expanded NKT cells. We hypothesized that selective proliferation of an NK1.1– precursor population that has recently emigrated from the thymus results in the expansion of a significant pool of NK1.1– NKT cells. However, we found that NKT cell dynamics in thymectomized and sham-thymectomized mice are indistinguishable, and that NK1.1– and NK1.1+ NKT cells respond with similar kinetics to in vitro stimulation with α-GalCer. As an alternative possibility, we considered that NKT cells down-regulate the NK1.1 marker for an extended time period after in vivo activation, as previously shown in vitro (31). Our findings show that α-GalCer-activated NKT cells progressively down-regulate surface NK1.1, and that levels remain significantly suppressed 7 days after α-GalCer injection. We therefore conclude that lack of NK1.1 expression on expanded NKT cells results from its gradual down-regulation after in vivo activation. Akin to TCR down-regulation, NK1.1 down-regulation may rescue NKT cells from AICD, reduce the sensitivity of these cells to antigenic stimulation, and protect the host against extensive inflammation and tissue damage. Further, because engagement of NK1.1 on NKT cells promotes Th1 cytokine production by these cells (35), lack of NK1.1 expression may have functional consequences with regard to cytokine production by NKT cells on repeated stimulation.
Prior models of NKT cell activation have suggested that quickly after their apparent demise in the spleen and liver, cells are repopulated in these organs through homeostatic proliferation from an NKT cell reservoir in the bone marrow (4). Our findings do not invoke a role for NKT cell reservoirs outside of the spleen or liver for repopulation of NKT cells in these organs. Interestingly, when comparing NK1.1 expression levels on NKT cells in different organs of mice 48–72 h after α-GalCer injection, we found a significant percentage of NK1.1+ cells in the bone marrow (Fig. 8C) but not other organs. The latter finding may account for the designation of the bone marrow as a reservoir for repopulating NKT cells by other investigators (4).
Our studies illustrate that surface marker expression is not a reliable method for identifying NKT cells. Thus, some of the prior studies with NKT cells that have relied on NK1.1 and/or TCR expression for identification of these cells may require reinterpretation. Quantitation of rearranged invariant TCRα chains among DNA may be the most reliable method for identifying and quantifying these cells.
On the basis of our findings, we propose the following model for the response of NKT cells to in vivo activation. Quickly after injection of α-GalCer, NKT cells down-regulate surface TCR expression and produce large amounts of cytokines, which in turn activate a variety of other cell types, including NK cells, dendritic cells, B cells, and conventional T cells. Although a fraction of NKT cells activated in this manner undergoes AICD, a significant pool of NKT cells persists and undergoes robust proliferation, resulting in significant expansion of these cells 2–3 days after the initial α-GalCer injection. This expanded population is NK1.1– as a result of NK1.1 down-regulation. Homeostatic mechanisms then control the size of the NKT cell population, which remains slightly expanded 7 days after α-GalCer injection. This model of NKT cell activation is consistent with the in vitro behavior of these cells and with their capacity to remember prior encounters with α-GalCer in vivo (7, 17, 18). The NKT cell response to other stimuli such as anti-CD3 antibodies, IL-12, and certain infectious agents, may exhibit similar population dynamics. For example, our studies with Salmonella infection indicate that the apparent loss of NKT cells observed quickly after infection is due, at least in part, to the loss of NK1.1 expression by these cells. However, in the case of a lymphocytic choriomeningitis virus (LCMV) infection, NKT cell loss appears to be due to apoptosis, probably because LCMV directly infects these cells (9).
NKT cells are promising targets for the development of novel vaccine adjuvants and immunotherapeutics (3). In many of these therapeutic protocols, α-GalCer was administered multiple times, typically every other day or twice per week. These protocols were developed on a trial-and-error basis and, therefore, lack a scientific foundation. In addition, in a recent phase I trial, patients with solid tumors received weekly α-GalCer injections by the i.v. route, which resulted in the quick (within 24 h) loss of NKT cells. A better understanding of NKT cell population dynamics, as provided here, should prove valuable for effective targeting of the NKT cell population for therapeutic purposes.
In conclusion, our results demonstrate unexpected dynamics in the response of NKT cells to in vivo activation. Our findings warrant a revision of existing models of NKT cell activation, will guide future investigations on the role of NKT cells in various immune responses, and should aid in the rational design of NKT cell-based therapeutic protocols.
Supplementary Material
Acknowledgments
We thank Kirin Brewery for providing synthetic α-GalCer; Dr. A. Bendelac for Vα14 transgenic mice; J. Wei, T. Vincent, and M. Nadaf for technical assistance; and Drs. D. Godfrey and M. Taniguchi for sharing information before publication. This work was supported by National Institutes of Health Grants HL68744, AI50953, NS44044 (to L.V.K.), AI42284 (to S.J.), and AI43407 (to C.-R.W.); the Juvenile Diabetes Research Foundation International (to S.J.); and the Swedish Cancer Foundation (to M.J.W.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: α-GalCer, α-galactosylceramide; AICD, activation-induced cell death; CFSE, carboxyfluorescein diacetate succinimidyl ester; MFI, mean fluorescence intensity; NKT, natural killer T; TCR, T cell receptor.
References
- 1.Kronenberg, M. & Gapin, L. (2002) Nat. Rev. Immunol. 2, 557–568. [DOI] [PubMed] [Google Scholar]
- 2.Taniguchi, M., Harada, M., Kojo, S., Nakayama, T. & Wakao, H. (2003) Annu. Rev. Immunol. 21, 483–513. [DOI] [PubMed] [Google Scholar]
- 3.Wilson, M. T., Singh, A. K. & Van Kaer, L. (2002) Trends Mol. Med. 8, 225–231. [DOI] [PubMed] [Google Scholar]
- 4.Eberl, G. & MacDonald, H. R. (1998) Immunity 9, 345–353. [DOI] [PubMed] [Google Scholar]
- 5.Eberl, G. & MacDonald, H. R. (2000) Eur. J. Immunol. 30, 985–992. [DOI] [PubMed] [Google Scholar]
- 6.Matsuda, J. L., Naidenko, O. V., Gapin, L., Nakayama, T., Taniguchi, M., Wang, C. R., Koezuka, Y. & Kronenberg, M. (2000) J. Exp. Med. 192, 741–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fujii, S., Shimizu, K., Kronenberg, M. & Steinman, R. M. (2002) Nat. Immunol. 3, 867–874. [DOI] [PubMed] [Google Scholar]
- 8.Giaccone, G., Punt, C. J. A., Ando, Y., Ruijter, R., Nishi, N., Peters, M., von Blomberg, B. M. E., Scheper, R. J., van der Vliet, H. J. J., van den Eertwegh, A. J. M., et al. (2002) Clin. Cancer Res. 8, 3702–3709. [PubMed] [Google Scholar]
- 9.Hobbs, J. A., Cho, S., Roberts, T. J., Sriram, V., Zhang, J., Xu, M. & Brutkiewicz, R. R. (2001) J. Virol. 75, 10746–10754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Daniels, K. A., Devora, G., Lai, W. C., O'Donnell, C. L., Bennett, M. & Welsh, R. M. (2001) J. Exp. Med. 194, 29–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Emoto, M., Emoto, Y. & Kaufmann, S. H. (1995) Eur. J. Immunol. 25, 3321–3325. [DOI] [PubMed] [Google Scholar]
- 12.Kirby, A. C., Yrlid, U. & Wick, M. J. (2002) J. Immunol. 169, 4450–4459. [DOI] [PubMed] [Google Scholar]
- 13.Leite-de-Moraes, M. C., Herbelin, A., Gouarin, C., Koezuka, Y., Schneider, E. & Dy, M. (2000) J. Immunol. 165, 4367–4371. [DOI] [PubMed] [Google Scholar]
- 14.Kawano, T., Cui, J., Koezuka, Y., Toura, I., Kaneko, Y., Motoki, K., Ueno, H., Nakagawa, R., Sato, H., Kondo, E., Koseki, H. & Taniguchi, M. (1997) Science 278, 1626–1629. [DOI] [PubMed] [Google Scholar]
- 15.Motsinger, A., Haas, D. W., Stanic, A. K., Van Kaer, L., Joyce, S. & Unutmaz, D. (2002) J. Exp. Med. 195, 869–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cui, J., Watanabe, N., Kawano, T., Yamashita, M., Kamata, T., Shimizu, C., Kimura, M., Shimizu, E., Koike, J., Koseki, H., et al. (1999) J. Exp. Med. 190, 783–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Burdin, N., Brossay, L. & Kronenberg, M. (1999) Eur. J. Immunol. 29, 2014–2025. [DOI] [PubMed] [Google Scholar]
- 18.Singh, N., Hong, S., Scherer, D. C., Serizawa, I., Burdin, N., Kronenberg, M., Koezuka, Y. & Van Kaer, L. (1999) J. Immunol. 163, 2373–2377. [PubMed] [Google Scholar]
- 19.Bendelac, A., Hunziker, R. D. & Lantz, O. (1996) J. Exp. Med. 184, 1285–1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morita, M., Motoki, K., Akimoto, K., Natori, T., Sakai, T., Sawa, E., Yamaji, K., Koezuka, Y., Kobayashi, E. & Fukushima, H. (1995) J. Med. Chem. 38, 2176–2187. [DOI] [PubMed] [Google Scholar]
- 21.Stanic, A. K., De Silva, A. D., Park, J. J., Sriram, V., Ichikawa, S., Hirabyashi, Y., Hayakawa, K., Van Kaer, L., Brutkiewicz, R. R. & Joyce, S. (2003) Proc. Natl. Acad. Sci. USA 100, 1849–1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gapin, L., Matsuda, J. L., Surh, C. D. & Kronenberg, M. (2001) Nat. Immunol. 2, 971–978. [DOI] [PubMed] [Google Scholar]
- 23.Overbergh, L., Valckx, D., Waer, M. & Mathieu, C. (1999) Cytokine 11, 305–312. [DOI] [PubMed] [Google Scholar]
- 24.Benlagha, K., Weiss, A., Beavis, A., Teyton, L. & Bendelac, A. (2000) J. Exp. Med. 191, 1895–1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carnaud, C., Lee, D., Donnars, O., Park, S. H., Beavis, A., Koezuka, Y. & Bendelac, A. (1999) J. Immunol. 163, 4647–4650. [PubMed] [Google Scholar]
- 26.Schonrich, G., Kalinke, U., Momburg, F., Malissen, M., Schmitt-Verhulst, A. M., Malissen, B., Hammerling, G. J. & Arnold, B. (1991) Cell 65, 293–304. [DOI] [PubMed] [Google Scholar]
- 27.Cai, Z., Kishimoto, H., Brunmark, A., Jackson, M. R., Peterson, P. A. & Sprent, J. (1997) J. Exp. Med. 185, 641–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu, H., Rhodes, M., Wiest, D. L. & Vignali, D. A. (2000) Immunity 13, 665–675. [DOI] [PubMed] [Google Scholar]
- 29.Benlagha, K., Kyin, T., Beavis, A., Teyton, L. & Bendelac, A. (2002) Science 296, 553–555. [DOI] [PubMed] [Google Scholar]
- 30.Pellicci, D. G., Hammond, K. J., Uldrich, A. P., Baxter, A. G., Smyth, M. J. & Godfrey, D. I. (2002) J. Exp. Med. 195, 835–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen, H., Huang, H. & Paul, W. E. (1997) J. Immunol. 158, 5112–5119. [PubMed] [Google Scholar]
- 32.Takeda, K., Hayakawa, Y., Van Kaer, L., Matsuda, H., Yagita, H. & Okumura, K. (2000) Proc. Natl. Acad. Sci. USA 97, 5498–5503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Osman, Y., Kawamura, T., Naito, T., Takeda, K., Van Kaer, L., Okumura, K. & Abo, T. (2000) Eur. J. Immunol. 30, 1919–1928. [DOI] [PubMed] [Google Scholar]
- 34.Ito, K., Karasawa, M., Kawano, T., Akasaka, T., Koseki, H., Akutsu, Y., Kondo, E., Sekiya, S., Sekikawa, K., Harada, M., et al. (2000) Proc. Natl. Acad. Sci. USA 97, 740–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Arase, H., Arase, N. & Saito, T. (1996) J. Exp. Med. 183, 2391–2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.