Abstract
Human polymorphonuclear leukocytes (PMNs or neutrophils) are essential to the innate immune response against bacterial pathogens. Recent evidence suggests that PMN apoptosis facilitates resolution of inflammation during bacterial infection. Although progress has been made toward understanding apoptosis in neutrophils, very little is known about transcriptional regulation of this process during bacterial infection. To gain insight into the molecular processes that facilitate resolution of infection, we measured global changes in PMN gene expression during phagocytosis of a diverse group of bacterial pathogens. Genes encoding key effectors of apoptosis were up-regulated, and receptors critical to innate immune function were down-regulated during apoptosis induced by phagocytosis of Burkholderia cepacia, Borrelia hermsii, Listeria monocytogenes, Staphylococcus aureus, and Streptococcus pyogenes. Importantly, we identified genes that comprise a common apoptosis differentiation program in human PMNs after phagocytosis of pathogenic bacteria. Unexpectedly, phagocytosis of Str. pyogenes induced changes in neutrophil gene expression not observed with other pathogens tested, including down-regulation of 21 genes involved in responses to IFN. Compared with other bacteria, PMN apoptosis was significantly accelerated by Str. pyogenes and was followed by necrosis. Thus, we hypothesize that there are two fundamental outcomes for the interaction of bacterial pathogens with neutrophils: (i) phagocytosis of bacteria induces an apoptosis differentiation program in human PMNs that contributes to resolution of bacterial infection, or (ii) phagocytosis of microorganisms such as Str. pyogenes alters the apoptosis differentiation program in neutrophils, resulting in pathogen survival and disease.
Human bacterial pathogens are exposed to a rapid and profound innate immune response characterized by recruitment of polymorphonuclear leukocytes (PMNs or neutrophils) to sites of infection (1). After phagocytosis by human PMNs, microorganisms are destroyed by reactive oxygen species (ROS) and microbicidal products contained within granules (2). Inasmuch as deficiency in many antimicrobial neutrophil components leads to increased host susceptibility to infection (3), an enhanced understanding of molecular signaling pathways induced in PMNs during phagocytosis of bacterial pathogens is crucial for improving treatment and outcome of infectious disease.
The human innate immune system initiates acute inflammation at the onset of infection. Although the inflammatory response is highly beneficial to the host, subsequent termination of infection-induced inflammation is critical for limiting tissue damage (4). To this end, neutrophil apoptosis likely facilitates resolution of inflammation caused by PMN activation. Thus, it follows that some human pathogens might modulate PMN apoptosis to cause disease, an idea supported by the finding that Anaplasma phagocytophilum delays neutrophil apoptosis to survive (5). Although the molecular basis for this phenomenon is unknown, recent studies have shown that bacteria are capable of globally modulating innate host inflammatory responses at the level of gene transcription in peripheral blood mononuclear leukocytes (6) and macrophages (7). Whereas macrophages are important in mediating chronic inflammation, neutrophils are essential in initiation and execution of the acute inflammatory response and subsequent resolution of bacterial infection. Consistent with this role, PMNs are the predominant immune cell in the acute stage of most bacterial infections. The global transcription-regulated processes that occur after phagocytosis of bacterial pathogens have not been elucidated, and relatively little is known about transcription networks that contribute to resolution of infection. To address this deficiency in knowledge, we studied global gene expression in human neutrophils after phagocytosis of a widely diverse group of pathogenic bacteria.
Materials and Methods
Detailed protocols are published as Supporting Methods, which is published on the PNAS web site, www.pnas.org.
Isolation of Human PMNs. Human PMNs were isolated from venous blood (8) of healthy donors in accordance with a protocol approved by the Institutional Review Board for Human Subjects, National Institute of Allergy and Infectious Diseases.
Bacterial Strains and Culture Conditions. All bacterial strains used in this study were originally clinical isolates and cause a wide range of diseases in humans, including tick-borne relapsing fever (Borrelia hermsii, spirochete), pneumonia, cellulitis, and toxic shock syndrome (Staphylococcus aureus and Streptococcus pyogenes, Gram+ cocci), pharyngitis and necrotizing fasciitis (Str. pyogenes), meningitis (Listeria monocytogenes, Gram+ bacillus, and intracellular pathogen), and pneumonia in patients with cystic fibrosis or chronic granulomatous disease (Burkholderia cepacia, Gram– bacillus). Strains were grown to early exponential phase for all experiments as follows: Bor. hermsii strain DAH, BSK-H complete medium, supplemented with 6.0% normal rabbit serum; Sta. aureus strain COL, tryptic soy broth; serotype M1 Str. pyogenes strain MGAS5005, Todd-Hewitt broth supplemented with yeast extract; L. monocytogenes strain ATCC 7644, brain heart infusion medium; and Bkl. cepacia strain 4A, Luria–Bertani broth.
Phagocytosis Experiments and RNA Preparation/Gene Expression Analysis. Phagocytosis and killing of bacteria by human PMNs and production of ROS were determined by using previously reported methods (9, 10). For gene expression experiments, phagocytosis assays were performed with 107 human PMNs and 108 bacteria (10:1 bacteria-to-PMN ratio) in wells of a 12-well tissue culture plate. At the indicated time points, cells were lysed directly with RLT buffer (Qiagen, Valencia, CA). Purified RNA was used to prepare labeled cRNA target (20 μg) for analysis on GeneChip Hu95A arrays (Affymetrix, Santa Clara, CA) according to standard GeneChip protocols (www.affymetrix.com/support/downloads/manuals/expression_s2_manual.pdf). Experiments were performed with PMNs from three separate individuals. Detailed information regarding data analysis is published in Supporting Methods. TaqMan analysis of gene expression was performed with conditions used for the microarray analysis by using an ABI 7700 thermocycler (Applied Biosystems) as described (11). TaqMan RNA samples were prepared identical to those for microarray analysis with the single exception of treatment with the RNase-free DNase set (Qiagen).
CXCR1 and CXCR2 Surface Expression. After phagocytosis of bacteria, PMN surface expression of IL-8 receptors was measured with flow cytometry by using antibodies specific for CXCR1 (5A12) and CXCR2 (6C6) (BD Biosciences, San Diego). All samples were analyzed by using a FACSCalibur flow cytometer (BD Biosciences).
PMN Apoptosis and Cell Lysis. DNA fragmentation in PMNs after phagocytosis was determined with an Apo-BRDU Apoptosis Detection Kit (BD Biosciences). Apoptosis and cell lysis were determined by morphological analysis using conventional light microscopy (12).
Results and Discussion
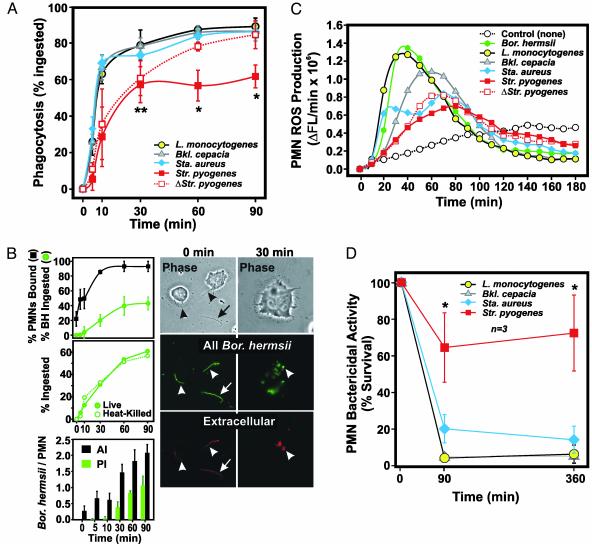
Phagocytosis and Killing of Bacterial Pathogens by Human PMNs. The ability of PMNs to kill a diverse array of bacterial pathogens is essential for human innate host defense. Therefore, we investigated phagocytosis of Bor. hermsii, L. monocytogenes, Bkl. cepacia, Sta. aureus, and Str. pyogenes by human neutrophils (Fig. 1). Phagocytosis of each pathogen occurred rapidly (within 10 min) and was virtually complete by 60 min, with one exception (Fig. 1 A). Compared with the other human pathogens, Str. pyogenes was significantly more resistant to phagocytosis at later times (P < 0.001 vs. other pathogens) (Fig. 1 A). This finding is consistent with the ability of Str. pyogenes to actively produce factors that modulate PMN function (9). Although we found that only 42.9 ± 8.5% of Bor. hermsii were completely ingested by 90 min in our assay (Fig. 1B), PMN binding (93.0 ± 7.0%) was similar to the other bacterial pathogens except Str. pyogenes (compare Fig. 1 A and B Top). Moreover, there was no difference in phagocytosis of live and dead Bor. hermsii (Fig. 1B Middle). Thus, although most Bor. hermsii were bound and at least partially ingested by PMNs, the unusual size (up to 20 μM in length) and shape of Bor. hermsii likely precluded complete phagosomal enclosure, resulting in low scoring of partially ingested organisms (Fig. 1B Right). Consistent with this idea, ROS production was greatest in PMNs stimulated with Bor. hermsii (Fig. 1C). In general, bactericidal activity correlated with phagocytosis and the magnitude of ROS produced by neutrophils (Fig. 1 C and D). Most notably, phagocytosis and ROS production were weakest in neutrophils stimulated with Str. pyogenes, a phenomenon that correlated well with increased pathogen survival.
Fig. 1.
Interaction of bacterial pathogens with human PMNs. (A) Phagocytosis of L. monocytogenes, Bkl. cepacia, Sta. aureus, and Str. pyogenes by human neutrophils. *, P < 0.001 vs. all bacteria; **, P ≤ 0.04 vs. all bacteria except heat-killed (Δ) Str. pyogenes. Results are the mean ± SD of three to five experiments. (B) Phagocytosis of Bor. hermsii.(Top) Number of PMNs with bound (black square) and/or ingested (green circle) Bor. hermsii. AI, association index; PI, phagocytic index. Micrographs (Right) illustrate ingestion and degradation of Bor. hermsii over time despite the appearance of extracellular staining (Extracellular, red). All Bor. hermsii, ingested, bound and extracellular (green). (Magnifications: ×400, Left; ≈×800, Right.) (C) PMN ROS production. The rate of ROS production for each pathogen is the mean of three separate experiments. ΔFL, change in fluorescence. (D) Killing of pathogens by human PMNs. At each time, PMNs were lysed and bacteria were plated on growth agar. Colonies were enumerated the next day, and percent bacteria killed was calculated as described in Supporting Methods. Results are the mean ± SD of three separate experiments. Bor. hermsii does not reliably form colony-forming units, and, thus, it was not possible to accurately assess PMN killing.
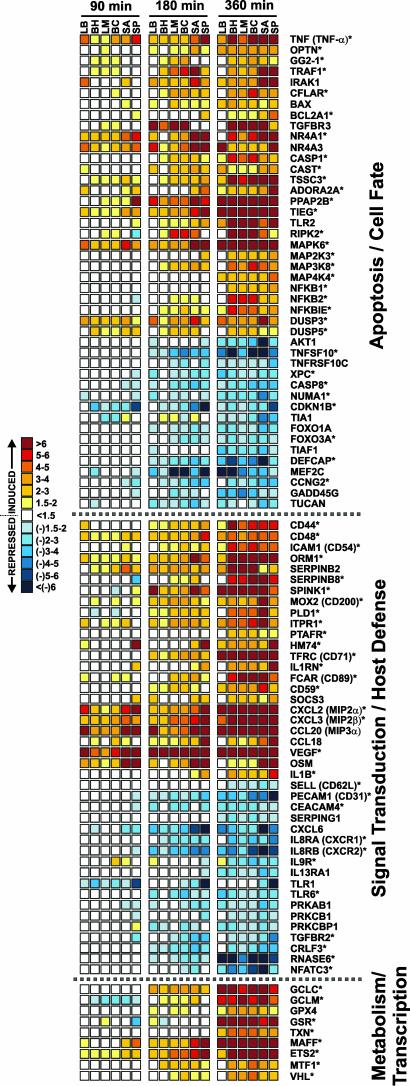
Bacterial Pathogens Induce Global Changes in PMN Gene Expression. Neutrophil function is driven by complex signaling pathways that involve generating and receiving diverse signals from a multiplicity of sources, including other host cells and bacterial pathogens. To gain insight into the molecular processes that are modulated by phagocytosis of bacterial pathogens in human neutrophils, we used oligonucleotide microarrays to analyze global changes in the PMN transcriptome after ingestion of Bor. hermsii, L. monocytogenes, Bkl. cepacia, Sta. aureus, and Str. pyogenes. We discovered a common pathogen-induced transcription profile that encompassed 305 up-regulated and 297 down-regulated genes over a period of 6 h after phagocytosis (Fig. 2, and Table 1, which is published as supporting information on the PNAS web site). Relatively few genes (81, ≈12%) were differentially expressed within 90 min after phagocytosis (Table 1). For example, only EGR1 and EGR2 were significantly upregulated at 20 min (Table 1). In contrast, 256 (43%) and 545 genes (91%) were differentially regulated 180 and 360 min, respectively, after phagocytosis of pathogens (Fig. 2 and Table 1). The finding that PMN gene regulation increases with time after phagocytosis of bacteria (3–6 h) is consistent with studies detailing receptor-mediated phagocytosis (10). A previous study reported only 27 common genes differentially expressed in neutrophils 2 h after exposure to Escherichia coli and Yersinia pestis (13), although the method used to measure gene expression was likely less sensitive.
Fig. 2.
A common pathogen-induced transcription profile in human PMNs. Shown are genes differentially expressed during PMN phagocytosis. Scale bar at left indicates fold change. Results are the mean fold-induction or repression of genes from three separate microarray experiments using three blood donors with phagocytosis assays done on separate days. *, P ≤ 0.05 vs. unstimulated PMNs. LB, IgG and C3bi-coated latex beads (for reference); BH, Bor. hermsii; LM, L. monocytogenes; BC, Bkl. cepacia; SA, Sta. aureus; SP, Str. pyogenes.
Transcriptional Regulation of Inflammatory Response Mediators. Phagocytosis of bacterial pathogens induces a molecular cascade of events in neutrophils that potentiate innate immune and inflammatory responses (2). At least 13 genes encoding proteins involved in activation and recruitment of immune effector cells were strongly induced by phagocytosis of pathogens, including CXCL2 [macrophage inf lammatory protein 2 (MIP-2α)], CXCL3 (MIP-2β), CCL20 (MIP-3α), vascular endothelial growth factor, and oncostatin M (Fig. 2 and Table 1). On the other hand, 82 genes encoding key surface molecules such as CXCR1, CXCR2, IL9R, TLR1, TLR6, CD31 (PEACAM1), IL13RA1, CD62L (L-selectin), and CEACAM4 were down-regulated. Down-regulation of CXCR1, CXCR2, CD31, and CD62L in PMNs after activation would diminish significantly their capacity for further chemotaxis and recruitment. The idea that IL-8 receptors are down-regulated after phagocytosis of pathogens is tested below. Taken together, these findings suggest that phagocytosis of bacteria down-regulates key inflammatory mediators several hours after stimulation, a time at which receptor-mediated phagocytosis has been shown to initiate PMN apoptosis. This observation is consistent with the discovery that there is down-regulation of proinflammatory capacity during programmed cell death in PMNs (14).
Genes Encoding Apoptosis Regulators Are Differentially Expressed After Phagocytosis of Bacterial Pathogens. A total of 105 apoptosis and cell fate-related genes were differentially transcribed after phagocytosis of bacterial pathogens, including genes encoding key mediators of tumor necrosis factor (TNF)-signaling such as TNF-α, GG2–1, TRAF1, TNFSF10 (TRAIL), TNFRSF10C (TRAILR3), and TSSC3 (Fig. 2). BAX and BCL2A1, two members of the BCL-2 family that play important roles in apoptosis in many types of eukaryotic cells, were up-regulated. The finding that BAX was up-regulated is consistent with our discovery that BAX is induced by ROS in human PMNs (S.D.K., J.M.V., K.R.B., A.R.W., W. M. Nauseef, H. L. Malech, and F.R.D., unpublished observations). Of note, genes encoding several members of the TLR2-signal transduction pathway, including TLR2, IRAK1, RIPK2, MAP4K4, CFLAR, IL1B, IL1RN, CASP1, NFKB1, and NFKB2 were up-regulated (Fig. 2). Activation of TLR2 by bacterial lipoprotein has been shown to induce programmed cell death in a variety of cell types including macrophages (15) and is independent of Fas and TNF (16). Importantly, our findings suggest that phagocytosis of bacterial pathogens modulates an apoptosis differentiation program in human PMNs that facilitates neutrophil programmed cell death.
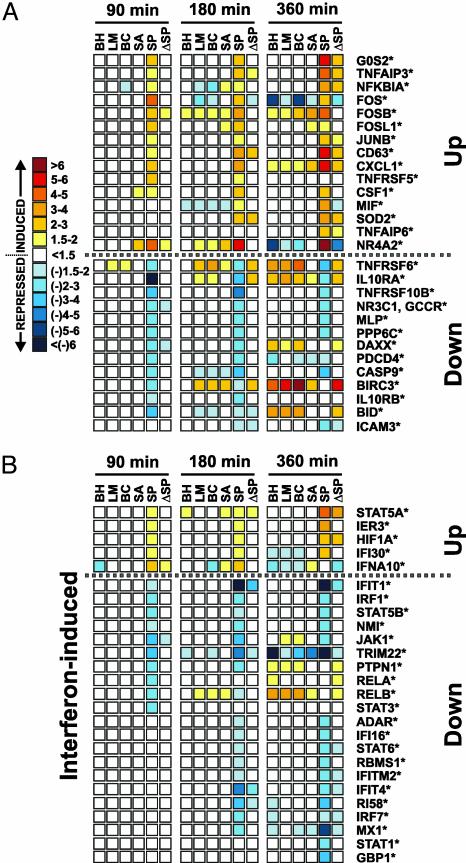
Str. pyogenes Alters the PMN Apoptosis Differentiation Program Induced by Other Pathogens. There were few pathogen-specific changes in neutrophil gene expression after phagocytosis of Bor. hermsii, L. monocytogenes, Bkl. cepacia, and Sta. aureus. By comparison, phagocytosis of Str. pyogenes resulted in up- or down-regulation of 393 genes (168 up-regulated and 225 down-regulated) whose expression differed significantly from that in PMNs stimulated with other bacteria (Fig. 3 and Table 2, which is published as supporting information on the PNAS web site). Of these genes, 197 (50%) were up- or down-regulated by 90 min, a much higher percentage than in the common program (12%) at the same time (compare Figs. 2 and 3). Notably, phagocytic interaction of Str. pyogenes with PMNs altered the expression of 71 apoptosis/cell fate-related genes, including up-regulation of activator protein-1 (AP-1) complex-related genes FOS, FOSL1, FOSB, JUNB, and TNFRSF5 (CD40) (Fig. 3A). The AP-1 complex is an important regulator of cell death (17). The gene encoding NFKBIA was up-regulated and those encoding RELA and RELB were down-regulated (Fig. 3A). These findings support the idea that NF-κB signal transduction is down-regulated early after phagocytic interaction with Str. pyogenes, a phenomenon that would favor decreased host cell survival. These results indicate that phagocytosis of Str. pyogenes alters the normal apoptosis differentiation program elicited by all bacterial pathogens (Fig. 2).
Fig. 3.
Str. pyogenes alters the PMN apoptosis differentiation program. (A) Str. pyogenes-specific modulation of apoptosis/cell fate-related genes in human PMNs. Symbols and scale bar are as described in the legend for Fig. 2. ΔSP, heat-killed Str. pyogenes. *, P ≤ 0.05 vs. unstimulated PMNs. (B) IFN-induced genes differentially expressed in PMNs after phagocytosis of Str. pyogenes.
Interferons have profound regulatory effects on the human immune system. Provocatively, phagocytosis of Str. pyogenes modulated expression of at least 26 genes involved in responses to IFN (18) (Fig. 3B). IFN-responsive genes IFIT1, IFIT4, IFITM2, IFI16, IRF1, MX1, and ADAR, and transcription factors that play key roles in IFN signaling and cell fate, including STAT1, STAT3, STAT5B, STAT6, JAK1, RELA, and RELB, were down-regulated (Fig. 3B and Table 2). These findings suggest that the JAK-STAT cell survival pathway is repressed (Fig. 3B). Consistent with that interpretation, we found that a JAK-STAT-regulated inhibitor of PMN apoptosis, BIRC3 (19), is down-regulated after phagocytosis of Str. pyogenes (Fig. 3B). It is crucial to note that the vast majority of Str. pyogenes-specific changes in neutrophil gene expression are mediated by live rather than heat-killed bacteria (Fig. 3). Taken together, these data strongly favor the idea that specific survival pathways are repressed in PMNs after phagocytosis of Str. pyogenes. Importantly, these results suggest that Str. pyogenes actively alters inflammation signaling networks and apoptosis in human PMNs at the level of gene transcription.
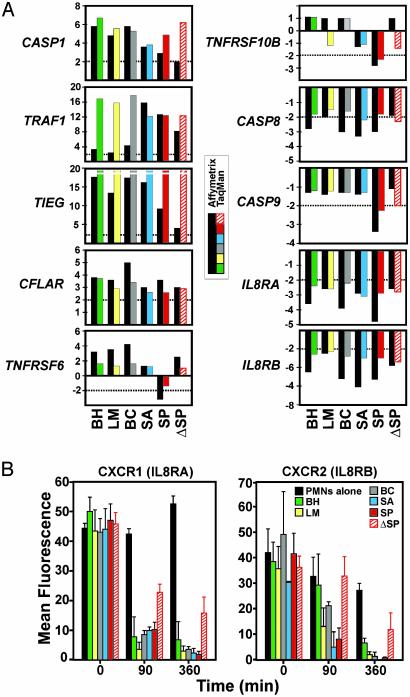
Confirmation of Microarray Data by TaqMan Real-Time RT-PCR and Flow Cytometry. We used TaqMan RT-PCR to verify changes in gene expression identified by microarray analysis (Fig. 4A). We selected 10 genes that encode key regulators of inflammation and apoptosis in neutrophils for confirmation. Eight of these genes are part of the common apoptosis differentiation program and two are changed only after phagocytosis of Str. pyogenes. There was strong positive correlation (≈90%) between oligonucleotide microarray data and TaqMan results (Fig. 4A), consistent with previous reports (10, 14).
Fig. 4.
Confirmation of microarray results. (A) Genes (n = 10) identified as differentially transcribed by microarrays were analyzed by TaqMan real-time PCR. TNFRSF6 and TNFRSF10B are representative of genes modulated only by phagocytosis of Str. pyogenes. There was a strong positive correlation (r = 0.90) between TaqMan and microarray results, consistent with previous comparisons (10, 14). Each bar represents the average change in PMN gene expression from three individuals assayed in triplicate. Abbreviations are as in the Fig. 2 legend. (B) Flow cytometric analysis of IL-8 receptors. After phagocytosis of the indicated bacteria, expression of CXCR1 (Left) and CXCR2 (Right) was measured by flow cytometry. Results are the net mean fluorescence ± SD of two phagocytosis experiments.
We next used flow cytometry to determine whether changes in surface expression of CXCR1 and CXCR2 reflected changes in gene expression (Fig. 4B). Phagocytosis of all bacteria resulted in decreased surface expression of CXCR1 and CXCR2, which was accompanied by down-regulation of the corresponding genes at 3 and6h(compare Figs. 2 and 4). Inasmuch as previous studies indicate that phagocytosis per se reduces surface expression of both receptors (20), it is likely that down-regulation of CXCR1 and CXCR2 early after phagocytosis of pathogens (within 90 min) is not regulated at the level of gene expression, an idea supported by our findings (see gene expression at 90 min). However, significantly reduced expression of genes encoding CXCR1 and CXCR2 at much later times after phagocytosis (e.g., at 3 and 6 h) likely contributes to the continued absence of expression of these receptors on the surface of human PMNs (Fig. 4B).
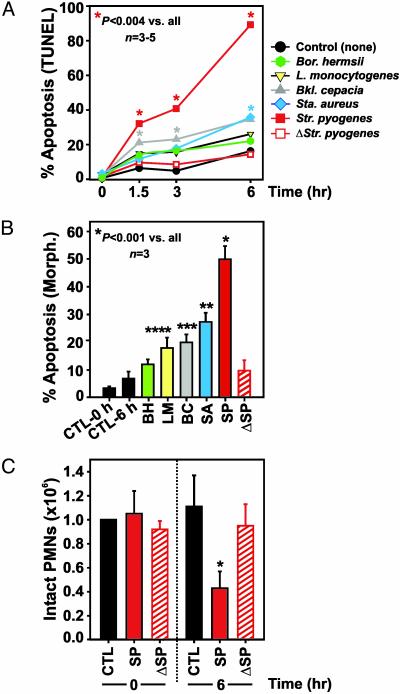
PMN Apoptosis Is Modulated by Bacterial Pathogens. Inasmuch as the common gene expression program suggests that phagocytosis of microorganisms induces programmed cell death in human neutrophils (Fig. 2), we measured PMN apoptosis after phagocantly cytosis of each bacterial pathogen (Fig. 5). Consistent with the microarray data, phagocytosis of all pathogens accelerated PMN apoptosis (Fig. 5). Unexpectedly, Str. pyogenes induced rapid apoptosis in neutrophils, which was significantly greater in magnitude than that elicited by the other pathogens (P ≤ 0.004). In fact, Str. pyogenes-induced PMN apoptosis at 90 min was similar to or exceeded levels induced by the other bacteria at 6 h (Fig. 5 A and B). The finding that heat-killed Str. pyogenes had little or no capacity for inducing PMN apoptosis is consistent with the idea that live Str. pyogenes produces factors that alter apoptosis in human PMNs (Fig. 5). Phagocytosis or ROS production per se were not independent predictors of pathogen-induced apoptosis, because ingestion and subsequent production of ROS were lowest in PMNs activated by Str. pyogenes (Fig. 1). Although phagocytosis and ROS have been reported to induce apoptosis in PMNs (10, 21), it is possible that minimal levels of ROS are necessary, but not sufficient for initiation of apoptosis (22). Live Str. pyogenes induced significant PMN necrosis not observed with phagocytosis of heat-killed organisms (Fig. 5C). Further, PMN necrosis correlated well with the accelerated apoptosis induced by interaction with Str. pyogenes (in Fig. 5 compare A and B with C). These observations coupled with the microarray data provide strong support to the idea that repression of genes controlling PMN fate is a key factor in Str. pyogenes pathogenesis.
Fig. 5.
Bacterial pathogens induce PMN apoptosis. (A) PMN apoptosis was assessed with a modified terminal deoxynucleotidyltransferase-mediated dUTP nick end labeling assay after phagocytosis of pathogens. Red asterisk, P < 0.004 vs. unstimulated PMNs and cells stimulated with all other pathogens. Gray and blue asterisks, P < 0.03 vs. unstimulated PMNs. Results are the mean ± SD of three to five separate experiments. (B) PMN apoptosis was assessed by nuclear morphology (Morph.) after phagocytosis of pathogens. *, P ≤ 0.001 vs. unstimulated PMNs and cells stimulated with all other pathogens; **, P ≤ 0.003 vs. unstimulated PMNs and cells stimulated with Bor. hermsii, and heat-killed (Δ) Str. pyogenes. ***, P = 0.01 vs. unstimulated PMNs; ****, P = 0.04 vs. unstimulated PMNs. (C) Str. pyogenes induces PMN necrosis. PMN lysis was measured by microscopy at the indicated times after phagocytosis. *, P < 0.001 vs. unstimulated PMNs and heat-killed Str. pyogenes. Results are the mean ± SD of three to six separate experiments. Abbreviations are as in the Fig. 2 legend.
Conclusions
Several lines of evidence indicate that bacterial pathogens induce an apoptosis differentiation program in PMNs that is essential for resolution of infection. First, phagocytosis of bacteria, production of ROS, and killing of microorganisms were followed by global changes in PMN gene expression common to a broad range of bacterial pathogens. Production of ROS accompanying neutrophil phagocytosis evokes changes in neutrophil gene transcription that, in part, facilitate apoptosis and likely mediate resolution of inflammation (S.D.K., J.M.V., K.R.B., A.R.W., W. M. Nauseef, H. L. Malech, and F.R.D., unpublished observations). Second, genes encoding proapoptosis factors were up-regulated and genes encoding antiapoptosis proteins were down-regulated after phagocytosis of all pathogens by human PMNs. Consistent with this observation, phagocytosis of bacteria induced PMN apoptosis, which was accompanied by down-regulation of proinflammatory molecules and genes encoding proinflammatory molecules.
Another important discovery in our study was that Str. pyogenes alters the apoptosis program in human neutrophils to survive, a process that has broad implications for infections caused by this pathogen. Of the pathogens used in this study, Str. pyogenes arguably causes the greatest number of human infections in “nonimmunocompromised” individuals, a phenomenon reflected by our in vitro data (see Fig. 1D). Microarray analysis was key to establishing a global model of host cell–pathogen interaction that provides fundamental insight into the resolution of infection in humans. Insight into bacterial evasion strategies revealed by our genomic studies of neutrophil function is likely applicable to other types of pathogenic microorganisms. Importantly, the pathogen-induced PMN apoptosis differentiation program revealed by our studies identified potential targets for therapeutic augmentation of host innate immunity during bacterial infections.
Supplementary Material
Acknowledgments
We thank S. Holland for Bkl. cepacia strain 4A and M. Schrumpf for assistance with growth of Bor. hermsii.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: PMN, polymorphonuclear leukocyte; ROS, reactive oxygen species; TNF, tumor necrosis factor.
References
- 1.Kaplanski, G., Marin, V., Montero-Julian, F., Mantovani, A. & Farnarier, C. (2003) Trends Immunol. 24, 25–29. [DOI] [PubMed] [Google Scholar]
- 2.Nauseef, W. M. & Clark, R. A. (2000) in Basic Principles in the Diagnosis and Management of Infectious Diseases, eds. Mandel, G. L., Bennett, J. E. & Dolin, R. (Churchill Livingstone, New York), Vol. 1, pp. 89–112. [Google Scholar]
- 3.Lekstrom-Himes, J. A. & Gallin, J. I. (2000) N. Engl. J. Med. 343, 1703–1714. [DOI] [PubMed] [Google Scholar]
- 4.Nathan, C. (2002) Nature 420, 846–852. [DOI] [PubMed] [Google Scholar]
- 5.Scaife, H., Woldehiwet, Z., Hart, C. A. & Edwards, S. W. (2003) Infect. Immun. 71, 1995–2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boldrick, J. C., Alizadeh, A. A., Diehn, M., Dudoit, S., Liu, C. L., Belcher, C. E., Botstein, D., Staudt, L. M., Brown, P. O. & Relman, D. A. (2002) Proc. Natl. Acad. Sci. USA 99, 972–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nau, G. J., Richmond, J. F., Schlesinger, A., Jennings, E. G., Lander, E. S. & Young, R. A. (2002) Proc. Natl. Acad. Sci. USA 99, 1503–1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boyum, A. (1968) Scand. J. Clin. Lab. Invest. Suppl. 97, 77–89. [PubMed] [Google Scholar]
- 9.Voyich, J. M., Sturdevant, D. E., Braughton, K. R., Kobayashi, S. D., Lei, B., Virtaneva, K., Dorward, D. W., Musser, J. M. & DeLeo, F. R. (2003) Proc. Natl. Acad. Sci. USA 100, 1996–2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kobayashi, S. D., Voyich, J. M., Buhl, C. L., Stahl, R. M. & DeLeo, F. R. (2002) Proc. Natl. Acad. Sci. USA 99, 6901–6907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chaussee, M. S., Watson, R. O., Smoot, J. C. & Musser, J. M. (2001) Infect. Immun. 69, 822–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Savill, J. S., Wyllie, A. H., Henson, J. E., Walport, M. J., Henson, P. M. & Haslett, C. (1989) J. Clin. Invest. 83, 865–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Subrahmanyam, Y. V. B. K., Yamaga, S., Prashar, Y., Lee, H. H., Hoe, N. P., Kluger, Y., Gerstein, M., Goguen, J. D., Newburger, P. E. & Weissman, S. M. (2001) Blood 97, 2457–2468. [DOI] [PubMed] [Google Scholar]
- 14.Kobayashi, S. D., Voyich, J. M., Braughton, K. & DeLeo, F. R. (2003) J. Immunol. 170, 3357–3368. [DOI] [PubMed] [Google Scholar]
- 15.Aliprantis, A. O., Yang, R. B., Mark, M. R., Suggett, S., Devaux, B., Radolf, J. D., Klimpel, G. R., Godowski, P. & Zychlinsky, A. (1999) Science 285, 736–739. [DOI] [PubMed] [Google Scholar]
- 16.Aliprantis, A. O., Yang, R. B., Weiss, D. S., Godowski, P. & Zychlinsky, A. (2000) EMBO J. 19, 3325–3336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shaulian, E. & Karin, M. (2002) Nat. Cell Biol. 4, E131–E136. [DOI] [PubMed] [Google Scholar]
- 18.Der, S. D., Zhou, A., Williams, B. R. G. & Silverman, R. H. (1998) Proc. Natl. Acad. Sci. USA 95, 15623–15628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hasegawa, T., Suzuki, K., Sakamoto, C., Ohta, K., Nishiki, S., Hino, M., Tatsumi, N. & Kitagawa, S. (2003) Blood 101, 164–1171. [DOI] [PubMed] [Google Scholar]
- 20.Doroshenko, T., Chaly, Y., Savitskiy, V., Maslakova, O., Portyanko, A., Gorudko, I. & Voitenok, N. N. (2002) Blood 100, 2668–2671. [DOI] [PubMed] [Google Scholar]
- 21.Coxon, A., Rieu, P., Barkalow, F. J., Askari, S., Sharpe, A. H., von Andrian, U. H., Arnaout, M. A. & Mayadas, T. N. (1996) Immunity 5, 653–666. [DOI] [PubMed] [Google Scholar]
- 22.Zhang, B., Hirahashi, J., Cullere, X. & Mayadas, T. N. (2003) J. Biol. Chem. 278, 28443–28454. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.