Abstract
Hypogonadotropic hypogonadism is defined as a deficiency of the pituitary secretion of follicle-stimulating hormone and luteinizing hormone, which results in the impairment of pubertal maturation and of reproductive function. In the absence of pituitary or hypothalamic anatomical lesions and of anosmia (Kallmann syndrome), hypogonadotropic hypogonadism is referred to as isolated hypogonadotropic hypogonadism (IHH). A limited number of IHH cases are due to loss-of-function mutations of the gonadotropin-releasing hormone receptor. To identify additional gene defects leading to IHH, a large consanguineous family with five affected siblings and with a normal gonadotropin-releasing hormone receptor coding sequence was studied. Homozygosity whole-genome mapping allowed the localization of a new locus within the short arm of chromosome 19 (19p13). Sequencing of several genes localized within this region showed that all affected siblings of the family carried a homozygous deletion of 155 nucleotides in the GPR54 gene. This deletion encompassed the splicing acceptor site of intron 4–exon 5 junction and part of exon 5. The deletion was absent or present on only one allele in unaffected family members. GPR54 has been initially identified as an orphan G protein-coupled receptor with 40% homology to galanin receptors. Recently, a 54-aa peptide derived from the KiSS1 protein was identified as a ligand of GPR54. The present study shows that loss of function of GPR54 is a cause of IHH, and it identifies GPR54 and possibly KiSS1 protein-derived peptide as playing a major and previously unsuspected role in the physiology of the gonadotropic axis.
The integrity of the pituitary–gonadal axis allows normal sexual differentiation during fetal life and normal puberty and fertility (1–3). Hypogonadotropic hypogonadism is defined by a defect of gonadal functions manifested by impuberism, partial pubertal development or isolated infertility, caused by a deficiency of follicle-stimulating hormone and luteinizing hormone production. The biological hallmark is a decreased level of sex steroids associated with low or normal levels of follicle-stimulating hormone and luteinizing hormone. This profile can result from deficiencies in gonadotropin-releasing hormone (GnRH) production by the hypothalamus, in GnRH receptor function at the pituitary level, or in luteinizing hormone and follicle-stimulating hormone production by the pituitary. Hypogonadotropic hypogonadism can result from tumoral, surgical, or physical insults at the hypothalamic or pituitary level and is generally associated with other pituitary hormone deficiencies. Hypogonadotropic hypogonadism can be part of multiple pituitary hormone deficiencies associated with genetic defects of transcription factors such as PROP1, HESX1, LHX3, and LHX4 or with developmental abnormalities of the region, such as pituitary stalk interruption syndrome (4). Hypogonadotropic hypogonadism can be associated with anosmia (Kallmann syndrome) or apparently isolated without anosmia [isolated hypogonadotropic hypogonadism (IHH)] (5). Kallmann syndrome is itself heterogeneous with an X-linked form due to mutations in Kal1, the gene encoding for anosmin (6, 7), and forms with autosomal transmission. Loss-of-function mutations of the fibroblast growth factor receptor 1 have recently been associated with ≈10% of autosomal forms of Kallmann syndrome (8). In 1997, the first cases of IHH due to loss-of-function mutations of the GnRH receptor gene were reported by our group (9). However, GnRH receptor mutations account for <50% of familial IHH cases and a small fraction of sporadic cases, indicating that IHH is a heterogeneous condition caused by various genetic defects (10).
In this study, a large consanguineous family comprising five siblings with IHH led us to identify, by a genome-mapping strategy, the G protein-coupled receptor 54 (GPR54) as a protein involved in the regulation of gonadotropin secretion.
Subjects, Materials, and Methods
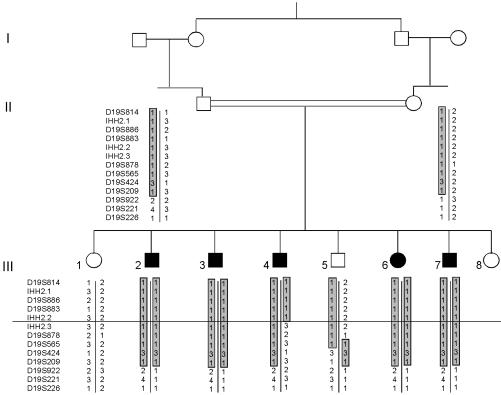
Subjects. A consanguineous family of eight children of whom five were affected was investigated (Fig. 1). The parents were first cousins. The index case was a 20-year-old male referred for impuberism. He had typical signs of hypogonadism with small testes (4 ml), sparse pubic hair (P3), and a penis of 7 cm. His height and weight were 152 cm and 54 kg, respectively, and his bone age was retarded at 15.0 years. He had a normal sense of smell and showed no abnormal eye movements, no color blindness, and no renal or craniofacial abnormalities. Three brothers had similar clinical features. A sister had partial hypogonadism; at age 16 years she had partial breast development and had experienced a single episode of uterine bleeding. Hormonal measurements (Table 1) showed low plasma testosterone in affected males and low plasma estradiol in the affected female, associated with low plasma gonadotropin levels. All affected siblings had a blunted response to GnRH (100 μg i.v.). One brother and two sisters had normal pubertal development. The mother had menarche at the age of 16 years. The father reported normal pubertal development. All patients and family members gave written informed consent for genetic analyses.
Fig. 1.
Haplotypes. The common haplotype shared by both parents is shown in gray boxes. IHH2.1, IHH2.2, and IHH2.3 are new CA repeat markers selected in the region of interest. A horizontal line shows the location of the recombination event in patient III.4.
Table 1. Hormonal status of affected siblings.
| GnRH test
|
||||||||
|---|---|---|---|---|---|---|---|---|
| Subjects (gender | Age, yr | Bone age, yr | Plasma testosterone, ng/dl | Plasma estradiol, pg/ml | Plasma LH, milliunits/ml | Plasma FSH, milliunits/ml | LH | FSH |
| III.2 (m) | 21 | 15 | 26 | - | 1.5 | 0.5 | 3.6 | 1.7 |
| III.3 (m) | 20 | 15 | 19 | - | 1.5 | 0.5 | 1.4 | 1.5 |
| III.4 (m) | 19 | - | 5 | - | 1.1 | 4.1 | 1.9 | 4.1 |
| III.6 (f) | 18 | - | - | 17 | 2.0 | 3.4 | 11.8 | 6.4 |
| III.7 (m) | 14 | 11 | 5 | - | 2.6 | 1.8 | 3.4 | 2.6 |
The chronological and bone ages are indicated. Normal values: males, luteinizing hormone (LH), 1.0-5.0 units/ml; follicle-stimulating hormone (FSH), 0.9-5.7 units/ml; testosterone, 260-690 ng/dl. Females, LH, 1.1-5.4 units/ml; FSH, 2.3-6.0 units/ml; estradiol (early follicular phase), 25-90 pg/ml. The GnRH test was performed by intravenous administration of 100 μg of GnRH. The highest values observed for plasma LH and FSH are reported. m, male; f, female.
Genotyping. Genomic DNA was isolated from peripheral lymphocytes by using standard methods. Genotyping was conducted at the Genopole in Evry, France. A genomewide analysis was performed by using the Genethon panel with microsatellite markers spaced, on average, ≈15 centimorgans apart (11). For fine mapping, we used other microsatellite markers published elsewhere (12) and new informative CA repeat markers (HH2.1, HH2.2, and HH2.3) that we identified from contigs localized within the region of interest. The primer sequences of these markers are listed in Table 2.
Table 2. Primer sequences of the new CA repeats used in the fine mapping.
| Primer name | Sequences | Genomic contig | Position of the 5′ nucleotide of the primer |
|---|---|---|---|
| IHH2.1F | CCAAGATCACACAACTGCAC | NT_011255.13 | 552 360 |
| IHH2.1R | ACCTGATAGTCTGACCGAAT | NT_011255.13 | 552 486 |
| IHH2.2F | CACGGAAGGGAAGCAACCAT | NT_011255.13 | 1 594 931 |
| IHH2.2R | TGATCTCACCACTGCACTCC | NT_011255.13 | 1 595 035 |
| IHH2.3F | ATAAATAAAGTGGCCCAGGA | NT_011255.13 | 1 718 816 |
| IHH2.3R | GGGGTCCGGAAACAGGTAG | NT_011255.13 | 1 718 933 |
Linkage Analysis. Two-point logarithm of odds (lod) scores were computed by using fastlink 4.1p (13, 14) and multipoint lod scores were computed by using genehunter 2.0 (15). The disease gene was assumed to be fully penetrant, autosomal, and recessive (frequency, 0.00001). Frequencies of the marker allele were varied to study the robustness of the results. All results are reported assuming equifrequent alleles (frequency, 0.25) at each marker. For multipoint analysis, genetic distances between markers were derived from the Genethon map (11).
Sequencing. The five exons of the GPR54 gene were amplified by PCR with 20–100 ng of genomic DNA. The following primers were used. Exon 1: forward, GGGCGGCCGGGAGGAGGA; reverse, CCGGGACGGCAGCAGGTG. Exon 2: forward, GCCCAGCGCCCGCGCATC; reverse, GTCCCCAAGTGCGCCCTCTC. Exon 3: forward, CAGGCTCCCAACCGCGCAG; reverse, CGTGTCCGCCTTCTCCCGTG. Exon 4: forward, CTTCATCCTGGCTTGTGGCAC; reverse, CTTGCTGTCCTCCCACCCAC. Exon 5: forward, GCCTTTCGTCTAACCACCTTC; reverse, GGAGCCGCTCGGA-TTCCCAC. Amplification was performed for 30 cycles with Yellow Taq (Eurogentec, Brussels) in 1.5 mM MgCl2, with 0.1 μM each primer and 5% DMSO. The annealing temperatures were 60°C for exons 1, 3, 4, and 5 and 66°C for exon 2. The PCR products were directly sequenced with BigDye dideoxyterminator cycle sequencing kits and the 3100 sequencer (Applied Biosystems) with the same primers. To genotype all members of the family, the PCR products of exon 5 were analyzed by electrophoresis in a 2% agarose gel.
Results
GnRH Receptor Genotyping. DNA was extracted from blood lymphocytes of the index case, and the GnRH receptor exons were sequenced and found to be normal. Two-point linkage analysis performed with markers localized on chromosome 4 allowed the definitive elimination of any anomaly of the GnRH receptor locus in this family (logarithm of odds score less than –2).
Genome Mapping. The large size of the family with numerous affected individuals and the high consanguinity allowed a homozygosity mapping strategy to localize the genomic region harboring the disease-causing allele. Preliminary computer analysis gave an 85% probability of finding the locus of interest (16) (data not shown). Two hundred eighty-two markers encompassing all autosomes were used for genome mapping (11). Only one marker, D19S886, was homozygous in all affected patients and was heterozygous in both parents. The logarithm of odds score for D19S886 was 3.5 at θ = 0, indicating a strong probability of linkage. A recombination event was found in patient III.4 between markers D19S886 and D19S424 (Fig. 1). D19S886 was the most telomeric marker used on the short arm of chromosome 19. The disease-causing genetic lesion was thus localized between the telomere (pTer) and D19S424. This region was called IHH2, spanning 14 centimorgans in genetic maps. We narrowed this interval by using four additional published markers (11, 12) and three previously undescribed polymorphic CA repeats (IHH2.1, IHH2.2, and IHH2.3) selected in the contigs localized within IHH2. Fine mapping using these markers assigned the recombination point to an interval of 110 kbp delimited by IHH2.2 and IHH2.3 (Fig. 2 Upper). The length of the interval of interest was estimated at 1,650 kbp (Homo sapiens Map View, Build 30; www.ncbi.nlm.nih.gov/mapview/maps.cgi).
Fig. 2.
(Upper) Localization and sequencing of the GPR54 gene. Markers used in the genotyping and the recombination point in patient III.4 are indicated. (Lower) DNA sequences from unaffected sib III.1 and affected patient III.2 are shown. The primers used for amplification of exon 5 were localized within intron 4 and in the 3′ untranslated region. The cDNA numbering was used to designate nucleotides in exon 5. In intron 4, nucleotides were numbered starting from the end of the intron. Residues of the 3′ end of intron 4 present in both individuals are underlined.
Sequencing of GPR54 Gene. Candidate genes present in the region of interest and potentially involved in reproduction, signaling, and development were sequenced. No difference was found between DNA from affected and nonaffected individuals in EPLG6, BSG, FSTL3, and NR3B genes (17–20). When amplifying exon 5 of the GPR54 gene of affected patients, we observed a single PCR product smaller than that of control subjects (Fig. 3). Exon 5 PCR amplification of DNA from both parents and from the unaffected sibling III.5 showed a band of the normal size and the smaller band. DNA from the unaffected sibling III.1 gave only the normal size band. A normal pattern of exon 5 PCR amplification was found in DNA from 50 control subjects. These results were consistent with the mapping data and suggested that a deletion within exon 5 of the GPR54 gene was linked to IHH in this family.
Fig. 3.
Genotyping of exon 5 in family. Exon 5 was amplified from genomic DNA. The PCR products (size in wild-type gene, 524 bp) were analyzed in 2% agarose gel.
Sequencing of the smaller PCR product amplified from affected patients showed a deletion of 155 bp lying between intron 4 (nucleotide –13 when numbering from the 3′ end of intron 4) and exon 5 (nucleotide 142 of exon 5, corresponding to nucleotide 880 of the cDNA) (Fig. 2 Lower). Sequencing of exon 5 in DNA amplified from the unaffected sibling III.5 confirmed the heterozygocity for the 155-bp deletion.
Discussion
In this paper, we show that a GPR54 gene defect leads to isolated hypogonadotropic hypogonadism. GPR54 is a G protein-coupled receptor initially cloned in the rat by using degenerate primers based on conserved sequences present in transmembrane domains (21). It has a 40% homology with galanin receptors. GPR54 was thereafter cloned in humans (called hOT7T175 or Axor-12) and found to be expressed mainly in the brain, pituitary, and placenta (22–24). GPR54 is formed of five exons. The deletion we have observed removes the splicing acceptor site of intron 4–exon 5 junction and part of exon 5 (Fig. 2 Upper). This deletion thus leads to the absence of the normal protein sequence downstream from residue 247. The deleted receptor is truncated within the third intracellular loop, thus lacking transmembrane domains 6 and 7. It has been shown that such truncated G protein-coupled receptors are unable to stimulate the transduction pathway (25). It is unknown whether the gene deletion present in this family leads to the absence of protein synthesis or to the synthesis of a truncated protein.
Recently, a ligand binding to GPR54 was described independently by three groups. Kotani et al. (22) and Ohtaki et al. (23) analyzed placental extracts for peptides activating GPR54. Muir et al. (24) used a library of 1,500 putative ligands. The best agonists displayed a similarity to a 54-aa peptide derived from the KiSS1 protein (26). This peptide corresponds to the predicted proteolytic processing of KiSS1 at dibasic and dibasic/amidation sites. It displays a C-terminal LRF-amide sequence. GPR54 stimulation by this 54-aa peptide results in the activation of phospholipase C by coupling to a Gq protein. KiSS1 mRNA is mainly present in the placenta and brain (24). In the brain, it has been localized to the hypothalamus and basal ganglia.
In support of our findings, we have sequenced the GPR54 gene in three other familial cases of isolated hypogonadotropic hypogonadism and found one missence homozygous point mutation (Leu102Pro). However, in contrast to deletions, point mutations must be expressed in transfected cells and shown to functionally impair the receptor. The Leu102Pro mutation in the GPR54 gene was not observed in 50 control subjects. Further studies of familial and sporadic cases of IHH will be necessary before assessing the frequency of GPR54 gene defects.
GPR54, initially described as an orphan receptor (21), binds a 54-aa RF-amide peptide derived by proteolysis from the KiSS1 protein (22–24). Our results show that GPR54 and possibly KiSS1-derived peptide play an important and previously unsuspected role in the physiology of the gonadotropic axis.
In conclusion, the present study shows that a genetic alteration leading to homozygous loss of function of GPR54 impairs pubertal development and reproductive functions. This finding indicates that GPR54 plays an important and previously unsuspected role in the regulation of the gonadotropic axis. The KiSS1 gene product is a RF-amide peptide, and two peptides with structural similarities have recently been shown to play a role in neuroendocrine regulations (27, 28). KiSS1 gene was initially cloned as a tumor metastasis suppressor gene (26), and the mature protein was named metastin. Activation of GPR54 by metastin results in decreased cellular motility (23) and proliferation (22, 23). Metastin is also believed to control the migration of trophoblast cells (29). However, it is unknown whether these features are relevant to the mechanisms leading to IHH in our patients. Moreover, further work is needed to determine whether GPR54 loss-of-function mutations affect the gonadotropic function at the hypothalamic or pituitary level. These findings suggest that a new chapter may thus be opened in the physiology of the gonadotropic axis.
Acknowledgments
We thank Odile Tinmar and Sylvain Goudet (Laboratoire d'Hormonologie et Biologie Moléculaire, Hôpital Bicêtre, Le Kremlin-Bicêtre, France) for their help with DNA sequencing, and Professors J. L. de Gennes and Paul Kelly for helpful discussions.
Abbreviations: IHH, isolated hypogonadotropic hypogonadism; GnRH, gonadotropin-releasing hormone; GPR54, G protein-coupled receptor 54.
References
- 1.Achermann, J. C., Ozisik, G., Meeks, J. J. & Jameson, J. L. (2002) J. Clin. Endocrinol. Metab. 87, 2447–2454. [DOI] [PubMed] [Google Scholar]
- 2.Themmen, A. P. N. & Huhtaniemi, I. T. (2000) Endocr. Rev. 21, 551–583. [DOI] [PubMed] [Google Scholar]
- 3.Kalantaridou, S. N. & Chrousos, G. P. (2002) J. Clin. Endocrinol. Metab. 87, 2481–2494. [DOI] [PubMed] [Google Scholar]
- 4.Cohen, L. E. & Radovick, S. (2002) Endocr. Rev. 23, 431–442. [DOI] [PubMed] [Google Scholar]
- 5.Seminara, S. B., Hayes, F. J. & Crowley, W. F., Jr. (1998) Endocr. Rev. 19, 521–539. [DOI] [PubMed] [Google Scholar]
- 6.Franco, B., Guioli, S., Pragliola, A., Incerti, B., Bardoni, B., Tonlorenzi, R., Carrozzo, R., Maestrini, E., Pieretti, M., Taillon-Miller, P., et al. (1991) Nature 353, 529–536. [DOI] [PubMed] [Google Scholar]
- 7.Legouis, R., Hardelin, J. P., Levilliers, J., Claverie, J. M., Compain, S., Wunderle, V., Millasseau, P., Le Paslier, D., Cohen, D., Caterina, D., et al. (1991) Cell 67, 423–435. [DOI] [PubMed] [Google Scholar]
- 8.Dode, C., Levilliers, J., Dupont, J. M., De Paepe, A., Le Du, N., Soussi-Yanicostas, N., Coimbra, R. S., Delmaghani, S., Compain-Nouaille, S., Baverel, F., et al. (2003) Nat. Genet. 33, 463–465. [DOI] [PubMed] [Google Scholar]
- 9.de Roux, N., Young, J., Misrahi, M., Genet, R., Chanson, P., Schaison, G. & Milgrom, E. (1997) N. Engl. J. Med. 337, 1597–1602. [DOI] [PubMed] [Google Scholar]
- 10.Beranova, M., Oliveira, L. M., Bedecarrats, G. Y., Schipani, E., Vallejo, M., Ammini, A. C., Quintos, J. B., Hall, J. E., Martin, K. A., Hayes, F. J., et al. (2001) J. Clin. Endocrinol. Metab. 86, 1580–1588. [DOI] [PubMed] [Google Scholar]
- 11.Dib, C., Faure, S., Fizames, C., Samson, D., Drouot, N., Vignal, A., Millasseau, P., Marc, S., Hazan, J., Seboun, E., et al. (1996) Nature 380, 152–154. [DOI] [PubMed] [Google Scholar]
- 12.Collin, G. B., Munch, A., Mu, J. L., Naggert, J. K., Olsen, A. S. & Nishina, P. M. (1996) Genomics 37, 125–130. [DOI] [PubMed] [Google Scholar]
- 13.Cottingham, R. W., Jr., Idury, R. M. & Schaffer, A. A. (1993) Am. J. Hum. Genet. 53, 252–263. [PMC free article] [PubMed] [Google Scholar]
- 14.Schaffer, A. A., Gupta, S. K., Shriram, K. & Cottingham, R. W., Jr. (1994) Hum. Hered. 44, 225–237. [DOI] [PubMed] [Google Scholar]
- 15.Kruglyak, L., Daly, M. J., Reeve-Daly, M. P. & Lander, E. S. (1996) Am. J. Hum. Genet. 58, 1347–1363. [PMC free article] [PubMed] [Google Scholar]
- 16.Genin, E., Todorov, A. A. & Clerget-Darpoux, F. (1998) Ann. Hum. Genet. 62, 419–429. [DOI] [PubMed] [Google Scholar]
- 17.Chatterton, J. E., Awobuluyi, M., Premkumar, L. S., Takahashi, H., Talantova, M., Shin, Y., Cui, J., Tu, S., Sevarino, K. A., Nakanishi, N., et al. (2002) Nature 415, 793–798. [DOI] [PubMed] [Google Scholar]
- 18.Drescher, U. (1997) Curr. Biol. 7, R799–R807. [DOI] [PubMed] [Google Scholar]
- 19.Fan, Q. W., Yuasa, S., Kuno, N., Senda, T., Kobayashi, M., Muramatsu, T. & Kadomatsu, K. (1998) Neurosci. Res. 30, 53–63. [DOI] [PubMed] [Google Scholar]
- 20.Guo, Q., Kumar, T. R., Woodruff, T., Hadsell, L. A., DeMayo, F. J. & Matzuk, M. M. (1998) Mol. Endocrinol. 12, 96–106. [DOI] [PubMed] [Google Scholar]
- 21.Lee, D. K., Nguyen, T., O'Neill, G. P., Cheng, R., Liu, Y., Howard, A. D., Coulombe, N., Tan, C. P., Tang-Nguyen, A. T., George, S. R., et al. (1999) FEBS Lett. 446, 103–107. [DOI] [PubMed] [Google Scholar]
- 22.Kotani, M., Detheux, M., Vandenbogaerde, A., Communi, D., Vanderwinden, J. M., Le Poul, E., Brezillon, S., Tyldesley, R., Suarez-Huerta, N., Vandeput, F., et al. (2001) J. Biol. Chem. 276, 34631–34636. [DOI] [PubMed] [Google Scholar]
- 23.Ohtaki, T., Shintani, Y., Honda, S., Matsumoto, H., Hori, A., Kanehashi, K., Terao, Y., Kumano, S., Takatsu, Y., Masuda, Y., et al. (2001) Nature 411, 613–617. [DOI] [PubMed] [Google Scholar]
- 24.Muir, A. I., Chamberlain, L., Elshourbagy, N. A., Michalovich, D., Moore, D. J., Calamari, A., Szekeres, P. G., Sarau, H. M., Chambers, J. K., Murdock, P., et al. (2001) J. Biol. Chem. 276, 28969–28975. [DOI] [PubMed] [Google Scholar]
- 25.Gether, U. (2000) Endocr. Rev. 21, 90–113. [DOI] [PubMed] [Google Scholar]
- 26.Lee, J. H., Miele, M. E., Hicks, D. J., Phillips, K. K., Trent, J. M., Weissman, B. E. & Welch, D. R. (1996) J. Natl. Cancer Inst. 88, 1731–1737. [DOI] [PubMed] [Google Scholar]
- 27.Satake, H., Hisada, M., Kawada, T., Minakata, H., Ukena, K. & Tsutsui, K. (2001) Biochem. J. 354, 379–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hinuma, S., Habata, Y., Fujii, R., Kawamata, Y., Hosoya, M., Fukusumi, S., Kitada, C., Masuo, Y., Asano, T., Matsumoto, H., et al. (1998) Nature 393, 272–276. [DOI] [PubMed] [Google Scholar]
- 29.Janneau, J. L., Maldonado-Estrada, J., Tachdjian, G., Miran, I., Motte, N., Saulnier, P., Sabourin, J. C., Cote, J. F., Simon, B., Frydman, R., et al. (2002) J. Clin. Endocrinol. Metab. 87, 5336–5339. [DOI] [PubMed] [Google Scholar]