Abstract
The Epstein–Barr virus (EBV) nuclear antigen 1 (EBNA1) is one of the earliest viral proteins expressed after infection and is the only latent protein consistently expressed in viral-associated tumors. EBNA1's crucial role in viral DNA replication, episomal maintenance, and partitioning is well examined whereas its importance for the immortalization process and the tumorgenicity of EBV is unclear. To address these open questions, we generated, based on the maxi-EBV system, an EBNA1-deficient EBV mutant and used this strain to infect primary human B cells. Surprisingly, lymphoblastoid cell lines (LCL) emerged from these experiments, although with very low frequency. These cell lines were indistinguishable from normal LCLs with respect to proliferation and growth conditions. A detailed analysis indicated that the entire viral DNA was integrated into the cellular genome. At least 5 of the 11 latent EBV proteins were expressed, indicating the integrity of the EBV genome. EBNA1-positive and ΔEBNA1-EBV-LCLs were injected into severe combined immunodeficient (SCID) mice to examine their tumorgenicity in comparison. Both groups supported tumor growth, indicating that EBNA1 is not mandatory for EBV's oncogenic potential. The results shown provide genetic evidence that EBNA1 is not essential to establish LCLs but promotes the efficiency of this process significantly.
Epstein–Barr virus (EBV) is a ubiquitous human herpesvirus that establishes latent infections and long-term persistence in human B lymphocytes. EBV is causally associated with various malignancies, including endemic Burkitt's lymphoma, nasopharyngeal carcinoma, Hodgkin's disease, and lymphoproliferative diseases in immunosuppressed patients (reviewed in ref. 1). In vitro EBV predominantly infects resting primary human B lymphocytes, which can acquire the characteristic of unlimited cell proliferation giving rise to lymphoblastoid cell lines, termed LCLs. This process partly reflects the pathogenetic mechanisms of tumor development and serves as a model for EBV's oncogenic capacity. In in vitro-infected and growth-transformed B cells, EBV establishes a latent infection in which only 11 of the 80 viral genes are expressed. Among them, five latent genes [the EBV nuclear antigens (EBNA) EBNA1, EBNA2, EBNA3A, and EBNA3C and the latent membrane protein 1 (LMP1)] are considered to be essential for this growth transformation (see ref. 2 for review).
In LCLs and EBV-associated tumors, the viral genome persists as a multicopy, extrachromosomal plasmid of ≈165 kbp in size. The EBV episome replicates once per cell cycle in synchrony with the host's genome and is maintained at a stable copy number in latently infected cells (for reviews, see refs. 3 and 4). Only two viral elements are required to ensure latent DNA replication and stable maintenance of the EBV genome. The latent origin of replication (oriP) is the cis-acting element, recognized by the virally encoded trans-acting EBNA1. EBNA1 is a dimeric, site-specific DNA-binding protein that binds to two elements within oriP, encompassing multiple cognate EBNA1-binding motifs (see ref. 5 for a recent review). Besides other latent viral genes, EBNA1 is detected in almost all types of latently infected cells and EBV-associated tumors. Moreover, it is the only viral protein exclusively expressed in type I latency characteristic of newly isolated Burkitt's lymphoma cells (1).
EBNA1 has a well-defined modular structure, including two arginine-rich basic domains that are involved in transcriptional activation, plasmid maintenance, and DNA replication (3, 6–9). Several cellular-interacting proteins have been identified that might be involved in plasmid maintenance, including the human p32/Tat-associated protein (10, 11), EBNA-1-binding protein 2 (12, 13), and Rch/importin α (14, 15). In addition, several interaction partners of EBNA1 have been identified that emphasize its function in DNA replication, such as the cellular origin recognition complex (ORC), other components of the prereplication complex (16–18), replication protein A (19), and the telomere repeat binding factor 2 (TRF2, ref. 20). EBNA1 has been shown to mediate random association of the EBV episomes and other oriP-containing plasmids with chromosomes of the cell, primarily during metaphase, indicating that EBNA1 plays a crucial role in EBV episome maintenance and partitioning to progeny nuclei (see ref. 5 for a recent review). These characteristics of EBNA1 might be brought about through its interaction with chromosomal proteins, such as high mobility group (HMG)-I and histone H1, which can functionally substitute for EBNA1's amino-terminal domain (21).
EBNA1 also acts as a bona fide transcriptional activator and enhances the expression of several viral genes from the viral Cp-promoter (22). EBNA1 was the first viral protein found to be expressed in EBV-associated human tumors and postulated to contribute to the growth-transformed state of EBV-infected cells directly (ref. 23 and references therein). Moreover, EBNA1 is associated with increased CD25 and V(D)J recombinase-activating genes RAG1 and -2 expression levels (24–26). These proteins might contribute to genetic instability and cellular transformation as suggested by EBNA1's possible oncogenic potential in transgenic mice (27–29). This view remains controversial because it has been deduced from gene expression that EBNA1 does not activate cellular target genes (23).
These various reports have led to the notion that EBNA1 is indispensable for in vitro growth transformation of primary human B lymphocytes (30), but this conviction has not been tested experimentally. Here, we prove that EBNA1 enhances the growth transformation of primary B cells by several thousandfold. The role of EBNA1 in our model system seems to support extrachromosomal replication because the novel ΔEBNA1-EBV-strain yields LCLs in which the viral genome is consistently found to be multiply integrated at variable sites of the host's genome.
Materials and Methods
Cells and Infection Experiments. Raji is a human EBV-positive Burkitt's lymphoma cell line; 293 cells are human embryonic epithelial kidney cells transformed by adenovirus; and Wi38 is a cell line of primary human embryonic fibroblasts. 293 cells were stably transfected with EBNA1 expressed from the cytomegalovirus promoter, and the EBV packaging cell line 293/TR– is already described (31). All cell lines and their cultivation are already described (31, 32). Primary human B lymphocytes were purified from human adenoids as described and infected with virus stocks (33). Infected B cells (1 × 104 to 1 × 105) were plated in a volume of 100 μl per well in 96-well cluster plates on Wi38 feeder cells and fed once a week. After 6–8 weeks, proliferating clones were expanded in suspension.
Recombinant EBV Plasmids. To generate the ΔEBNA1-EBV-strain, the entire EBNA1 ORF was replaced by insertional mutagenesis in Escherichia coli DH10B as described in detail (31, 32, 34). To this end, a conventional recombinant plasmid termed p2761.UA1 was constructed. A selectable marker encoding resistance against kanamycin was positioned in between nucleotide coordinates 107,949 and 109,873 of the B95.8 genome to precisely replace the entire ORF of EBNA1, only. For homologous recombination in E. coli, p2761.UA1 was cleaved with SspI and MluI (nucleotide coordinate 106,418 and 110,770 of the B95.8 genome, respectively) to provide the EBV-flanking regions for targeted insertional mutagenesis in E. coli DH10B carrying the maxi-EBV p2089 as described (35) to yield the ΔEBNA1-EBV-mutant p2828. DNAs from several single colonies were analyzed with restriction enzymes, and the genetic composition was confirmed by Southern blot hybridizations. One clone (no. 8) was selected, and large-scale plasmid DNA was prepared. The p2828 EBV clone 8 was transfected into 293 cells stably expressing EBNA1 in trans to permit stable EBV episome maintenance and replication. Single GFP-expressing and hygromycin-resistant cell clones that carry the EBV plasmid were selected for virus production on transient cotransfection of expression plasmids encoding BZLF1 and BALF4 as described (34, 35). The virus stocks 2828 were then used to incubate Raji cells to analyze the presence of infectious EBV, visualized indirectly by GFP expression as described (36).
For trans-complementation, the EBNA1 expression plasmid p2852 was constructed, in which EBNA1 is expressed as the only viral gene from the immediate-early enhancer and promoter of the human cytomegalovirus as shown in detail in Fig. 1C. Detailed cloning strategies and information about intermediate plasmids are available on request.
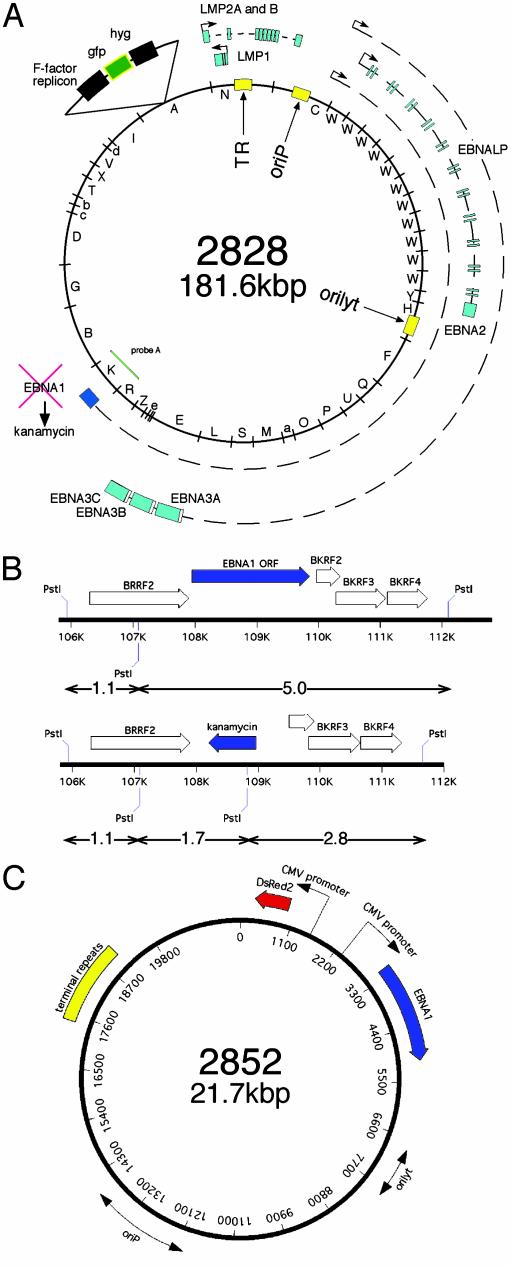
Fig. 1.
(A) Schematic picture of ΔEBNA1-genome p2828: The circular episome of the B95.8 strain is shown with its BamHI fragments (inner circle). Nine of the 11 latent viral genes (EBNA1, EBNA2, EBNA3A, -B, and -C, EBNA-LP; LMP1, LMP2A, and -2B) generally expressed in LCLs are depicted as blue boxes together with their primary transcripts and promoters. The EBNA1 gene was deleted by homologous recombination and replaced by the kanamycin gene. (B) Enlargement of the EBNA1-flanking regions used for homologous recombination to replace EBNA1 by kanamycin. The kanamycin resistance gene contains a diagnostic PstI restriction site used to distinguish the ΔEBNA1-genome p2828 (Lower) from the wild-type EBNA1 situation (Upper) by Southern blotting (Fig. 2B). (C) Composition of plasmid p2852 used for transcomplementation experiments in which both EBNA1 and the DsRed2 gene are expressed from the cytomegalovirus promoter. Both origins of DNA replication are indicated as double arrows, and the terminal repeats are shown as a yellow box.
Southern and Gardella Blot Hybridizations and Fluorescence in Situ Hybridization. Total cellular DNA extractions, restriction digests, and Southern blot hybridizations were performed according to standard protocols. Gardella gel electrophoresis and blotting procedures were done as described (37), followed by hybridization with 32P-labeled p135.16 plasmid DNA as probe (38). Fluorescence in situ hybridizations were performed essentially as described (39).
Western Blot analysis. For the detection of LMP2A, membrane fractions of cells were prepared in the presence of protease inhibitors and used for Western blot immunostainings with the rat monoclonal antibody 14B6 as described (40). A detailed protocol is available on request.
To detect EBNA1, -2, -3B, and -3C, and LMP1 by conventional immunostaining, the following primary antibodies were used: LMP1 (CS1-4 mouse, DAKO), EBNA1 (1H4 rat), EBNA2 (R3 rat), EBNA3B (5F6 rat), and EBNA3C (E3c.A10 mouse). Immunoblots were developed by using peroxidase-coupled secondary antibodies (Promega and Dianova, Hamburg, Germany) and the enhanced chemiluminescence system (ECL, Amersham Pharmacia).
Mice. Ten- to 12-week-old severe combined immunodeficient (SCID) mice were injected i.p. with 3 × 107 or 1 × 108 cells for each LCL in a volume of 300 μl of PBS as described (32). Tumor material, blood cells, and organ samples were analyzed for GFP expression and by Western blot immunostainings for EBNA2 and EBNA1.
Results
Construction and Production of ΔEBNA1-EBV 2828. The complete EBV genome of the prototype strain B95.8 was cloned in E. coli onto an F-factor plasmid to generate the wild-type-EBV p2089 (41). Genetic cloning of the entire genome resulted in a fully functional viral genome that could be readily manipulated genetically in the prokaryotic host. To substitute the EBNA1 coding region by the kanamycin gene, a linear DNA fragment was used for homologous recombination with the EBV plasmid p2089 as described (35). ΔEBNA1-EBV 2828 virions were produced by transfecting the p2828 genome into 293 cells stably expressing EBNA1 from an integrated copy. One cell clone was used throughout the following experiments that yielded up to 106 infectious particles per ml as does the wild-type strain 2089.
ΔEBNA1-EBV 2828 Transforms Primary Human B Cells at a Low Rate. To test whether ΔEBNA1-EBV 2828 induces sustained B cell proliferation, primary B cells were infected and cultivated for at least 6 wk on irradiated human fibroblast feeder layers (WI38) as described (42). Surprisingly, ΔEBNA1-EBV 2828 gave rise to growth-transformed, proliferating LCLs. From eight different B cell donors, 31 independent ΔEBNA1-LCLs were established and further expanded without feeder cells. It was immediately apparent that the efficiency of B cell growth transformation by the ΔEBNA1-EBV-strain was dramatically reduced compared with wild-type virus 2089. The very low immortalizing rate of the ΔEBNA1-EBV 2828 made it difficult to determine its growth transforming efficiency. At least 10,000-fold more 2828 virions were necessary to yield a proliferating ΔEBNA1 cell as in parallel experiments with wild-type EBV (32). The period needed to establish a B cell clone seemed to be slightly prolonged compared with wild-type EBV.
This phenotype could result either from the introduced deletion of the entire EBNA1 ORF or from accidental genetic alterations elsewhere in the ΔEBNA1 p2828 genome. To distinguish these scenarios, we constructed the EBV-based gene vector p2852 (Fig. 1C; refs. 31 and 43) that trans-complements EBNA1 from a heterologous promoter. p2852 does not encode for any other viral gene that might interfere with the immortalization process. This vector can be packaged into an EBV coat with the aid of the helper cell line TR– as described (31). B cell lines were established by coinfecting primary B cells with both the EBNA1 trans-complementing gene vector stock and the ΔEBNA1-EBV 2828. B cell lines established by coinfections were analyzed in parallel with B cell lines infected only with ΔEBNA1 2828 virus stocks.
To analyze the integrity of the growth-transformed B cells, Western blot immunostainings and Southern blot hybridizations were performed. In Fig. 2, B cell lines obtained from three different donors are shown. The B cell lines were obtained either with ΔEBNA1-EBV 2828 (Fig. 2, lanes 4–8) or from coinfection experiments with the trans-complementing EBNA1-vector (Fig. 2, lanes 9–12). As expected, lanes 9–12 illustrate EBNA1-specific signals in Western blot analysis similar to those of the lymphoblastoid B cell lines infected with wild-type EBV 2089 and the Raji cell line (Fig. 2 A). Lymphoblastoid B cells infected with ΔEBNA1-EBV 2828 in lanes 4–8 show no signal. Southern blot analysis of complete cellular DNA cleaved with PstI and hybridized to probe A (Fig. 1B) was performed to distinguish between the composition of ΔEBNA1 2828 and wild-type 2089 EBV (Fig. 2B). B cell lines infected with ΔEBNA1-EBV, alone or in combination with the EBNA1 trans-complementing gene vector, showed a pattern indicative of the deleted EBNA1 locus (Fig. 1B) whereas Raji cells and wild-type EBV infected B cells showed the pattern characteristic of generic EBV. These results provide strong genetic evidence that ΔEBNA1-EBV supports growth transformation of primary human B cells in vitro.
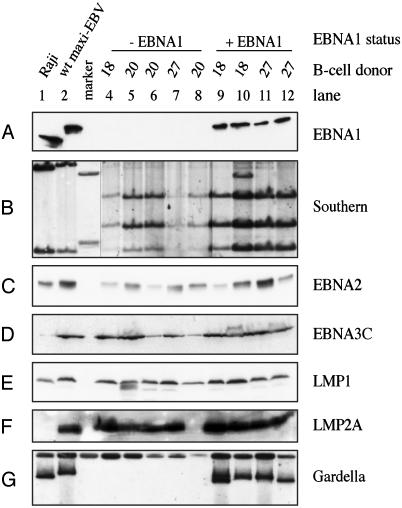
Fig. 2.
Analysis of LCLs generated by different EBV strains. ΔEBNA1-LCLs from three different donors (donors 18, 20, and 27) were obtained from infections of human primary B cells without (lanes 4–8) or with the EBNA1 trans-complementing plasmid p2852 (lanes 9–12). Raji cells (lane 1) and a generic EBV-LCL (lane 2) were used as controls. Raji is negative for EBNA3C and LMP2A due to deletions in the genome. (A) Western blot analysis of EBNA1. (B) Southern blot of genomic DNA to distinguish generic EBV from ΔEBNA1-LCLs. The sizes of diagnostic DNA fragments are shown in Fig. 1B, and the location of probe A is depicted in Fig. 1 A. Lanes 4–8 are from the same Southern blot as the other lanes, but were exposed longer. (C–F) Western blot analysis of different latent EBV proteins: EBNA2 (C), EBNA3C (D), LMP1 (E), and LMP2A (F) are expressed in all LCLs, with the exception of LMP2A (lane 8). (G) Gardella gel analysis used to determine the status of the viral DNA by using p135.16 as hybridization probe. EBV genomes are extrachromosomal when EBNA1 is present (lanes 1, 2, and 9–12) but integrated in the absence of the viral protein (lanes 4–8).
All Latent Genes of EBV Are Expressed in Growth-Transformed B Cells Established with ΔEBNA1-EBV 2828. Because EBNA1 transactivates several latent viral genes, it was interesting to learn whether other latent genes were expressed in LCLs established with ΔEBNA1-EBV 2828. Western blot analyses with different antibodies specific for latent EBV gene products were performed. All B cell lines in this study, including the control LCL infected with wild-type EBV 2089, were found to express EBNA2 (Fig. 2C), EBNA3B (data not shown), EBNA3C (Fig. 2D), LMP1 (Fig. 2E), and (with the exception of the cell line shown in lane 8) LMP2A (Fig. 2F). EBV-positive Raji cells do not express EBNA3C and LMP2A due to two internal deletions in the Raji EBV genome (44), but do express all other latent genes analyzed here. No obvious differences in signal intensities were detectable in these and other Western blot experiments. We expect that the remaining latent genes EBNA3A, EBNA-LP, and LMP2B are also expressed due to their common promoter usage. This finding means that all latent genes of EBV are likely consistently expressed in LCLs established by ΔEBNA1-EBV.
Extrachromosomal Viral DNAs Are Not Detectable in ΔEBNA1-EBV-Infected B Cells. All published data indicate that EBNA1 is obligatory for EBV's episomal DNA replication and maintenance. This observation contributes to the conviction that EBNA1 would also be indispensable for in vitro growth transformation of primary B cells. Because we have found EBNA1 not essential, we have tested whether EBV DNA is integrated into the host genome. We performed Gardella gel analysis to detect large extrachromosomal DNAs (ref. 37 and references therein). The episomal status of EBV genomes was proven by signals of LCLs expressing EBNA1 (Fig. 2G, lanes 1, 2, and 9–12; ref. 39) whereas B cell lines established with the ΔEBNA1-EBV 2828 only clearly lacked such a characteristic signal (Fig. 2G, lanes 4–8).
Multiple Chromosomal Integration of EBV Genomic DNA in ΔEBNA1-LCLs. To confirm chromosomal integration of EBV DNA in B cells infected with ΔEBNA1-EBV, we performed fluorescence in situ hybridization analysis on all B cell lines analyzed in Fig. 2. As an example, Fig. 3 shows four growth-transformed B cell lines of two different B cell donors corresponding to cells investigated in Fig. 2 [lane 7 (Fig. 3A), lane 4 (Fig. 3B), lane 11 (Fig. 3C), and lane 9 (Fig. 3D)]. The hybridization signals clearly show individual integration events of EBV DNA in metaphase chromosomes of cells infected with ΔEBNA1-EBV 2828 in contrast to multiple extrachromosomal EBV genomes loosely associated with the chromosomes in the presence of EBNA1. Metaphase chromosome hybridizations of the different B cell lines infected with ΔEBNA1-EBV suggest that EBV DNA could be found integrated randomly without any apparent chromosomal preference (data not shown). Our data also indicate that these B cell lines are likely monoclonal in origin, as reflected by the individual integration pattern of dual signals on sister chromatids (37, 39, 45).
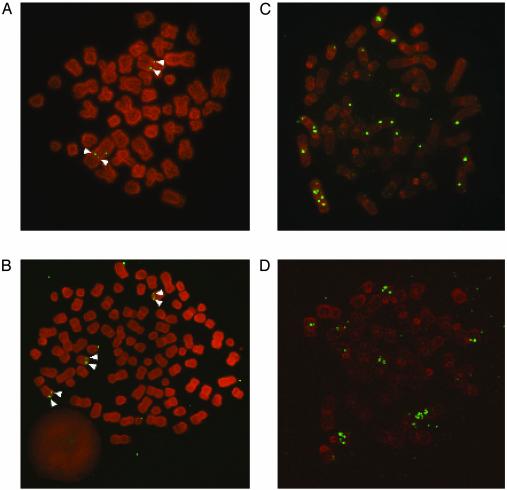
Fig. 3.
Metaphase chromosomes of LCLs were analyzed for the status of the viral genome by in situ hybridization. As representative examples, two different donors are shown: donor 27 (A and C) and donor 18 (B and D) (see Fig. 2). (A and B) ΔEBNA1-EBV 2828-derived LCLs show integration events of the viral genome into host chromosomes. Individual LCLs are of clonal nature as integration sites appear as doublets (white arrowheads). (C and D) ΔEBNA1-EBV-LCLs generated by coinfection with 2852 virus maintain the ΔEBNA1-EBV genome episomally with high copy number. Most viral episomes are randomly attached to metaphase chromosomes. A corresponds to lane 7 in Fig. 2, B to lane 4, C to lane 11, and D to lane 9.
Two copies of the EBV genome were integrated into the genome of donor 27, presumably in chromosome 1 and 3, as indicated by their characteristic shapes (Fig. 3A). As can be seen from the number of signals in Fig. 3, LCLs expressing EBNA1 display higher copy numbers of the EBV genome than LCLs in which EBV DNA is found chromosomally integrated. This finding is reminiscent of signal intensities observed in the Southern blot hybridizations shown in Fig. 2B. Southern blot hybridizations with the complete EBV DNA as a radioactive probe on total cellular DNA revealed that the overall restriction pattern characteristic of wild-type or ΔEBNA1-EBV-genomes was preserved even in cell lines where EBV was found integrated (data not shown). Polyploidy was observed in some of the cell lines analyzed (i.e., Fig. 3).
Our results support the hypothesis, that ΔEBNA1-EBV 2828 is able to growth transform primary human B cells in cases where the EBV genome successfully integrates into the host genome. Expression of all latent EBV genes (excluding EBNA1 and in one case LMP2A; Fig. 2F, lane 8) seemed to be a prerequisite to obtain such B cell lines. All cell lines generated in this study expressed typical surface activation markers characteristic of “classical” LCLs irrespective of the presence or absence of EBNA1 (data not shown).
The Oncogenic Potential of Growth-Transformed ΔEBNA1 B Cell Lines in SCID Mice. Human B cells infected with EBV cause lymphoproliferative malignancies in SCID mice (see ref. 46 for a recent review), which lack functional B and T cells and are incapable of rejecting xenografts. Human B cells infected with wild-type EBV are oncogenic in these mice and cause lymphoproliferative malignancies. Because EBNA1 was thought to be essential for the process of growth transformation of human B cells and for EBNA1 transgenic mice to develop tumors (27, 29), it was interesting to investigate the oncogenic potential of LCLs infected with ΔEBNA1-EBV 2828 in SCID mice.
Groups of mice were injected i.p. with 3 × 107 or 1 × 108 cells of two B cell donors. LCLs obtained after infection with wild-type EBV 2089, ΔEBNA1-EBV 2828, or ΔEBNA1-EBV trans-complemented with 2852 virus were used. Mice were killed when they became seriously ill and analyzed for evidence of tumor material. Biopsy material was used to reestablish injected human B cells, which were analyzed subsequently for the expression of EBNA1, EBNA2, and GFP. Growth-transformed B cells infected with either ΔEBNA1-EBV 2828, EBNA1 trans-complemented ΔEBNA1-EBV, or wild-type-EBV 2089 were able to proliferate in SCID mice. The injected B cells could be reestablished from the tumor mass and from blood samples of these animals as indicated by their expression of EBNA2 and GFP (data not shown).
Discussion
EBNA1 is the only latent protein that is found to be consistently expressed in EBV-associated tumor cells and is one of several viral genes that is considered to be essential for growth transformation of primary B cells (2). We therefore have analyzed EBNA1 genetically by infecting primary B cells in vitro with the ΔEBNA1-EBV-mutant 2828 (Fig. 1 A). We obtained genetically pure virus stocks with up to 1 × 106 particles per ml. Unexpectedly, growth-transformed LCLs could be established with ΔEBNA1-EBV 2828. However, the deletion of EBNA1 reduced the efficiency of growth transformation by at least 10,000-fold, as at least 10,000-fold more 2828 virions were required to generate a proliferating ΔEBNA1 cell. Analyzing the ΔEBNA1-LCLs indicated that the complete viral genome, including oriP, is integrated into the host genome. ΔEBNA1-EBV 2828 stocks can be trans-complemented by the EBNA1-expression vector p2852 (Fig. 1), which results in stable episomal maintenance of the ΔEBNA1-EBV 2828 genome in coinfected and growth-transformed B cell lines. All latent viral genes tested are expressed to comparable levels with or without EBNA1. These findings indicate that ΔEBNA1-EBV is, in contrast to ΔEBNA2- or ΔLMP1-EBV-strains, able to transform human B lymphocytes without obvious limitations. EBV mutants lacking EBNA2, EBNA-LP, or EBNA3C are completely unable to transform B cells (ref. 38 and unpublished data), whereas the ΔLMP1-EBV-strain depends on effects of fibroblast feeder cells (32). SCID mice were injected i.p. with ΔEBNA1-LCLs in parallel with B cell lines transformed either with EBNA1-trans-complemented ΔEBNA1-EBV or wild-type-EBV 2089 to address their oncogenic potential. Irrespective of their genetic composition, the B cell lines were able to proliferate in SCID mice and to yield malignant growth.
Several aspects are noteworthy: (i) in vitro growth transformation of primary B cells does not require EBNA1 although it improves the frequency of this process significantly; (ii) deletion of EBNA1 does not alter the profile of latent viral genes expressed in established LCLs; (iii) EBNA1 does not play a crucial role in the survival of SCID mice on LCL injection. We conclude from these data that growth transformation of primary human B cells by EBV per se is independent of EBNA1, which seems to contribute indirectly to this process. Very likely, EBNA1 guarantees extrachromosomal replication and genome maintenance to establish EBV's latent status efficiently. In the absence of EBNA1, the viral episome might be degraded shortly after infection in most primary B cells. In rare cases, the viral genome multiply integrates into the host's genome. An additional level of complexity might stem from the integrity of the viral genome to allow faithful expression of latent viral genes such as EBNA-LP, EBNA2, and EBNA3s from their genuine viral promoters Cp and Wp. Up to date, we do not know whether the deletion of EBNA1 has any in vivo relevance for the Cp promoter activity that is regulated by EBNA1 and/or EBNA2 via the FR-element within oriP (ref. 47 and references therein). It is even conceivable that the Wp to Cp promoter switch does not occur in the ΔEBNA1 strain, although this event is generally considered to be a critical step in the temporal regulation of latent viral genes (1). Currently, we cannot exclude that EBNA1 has multiple functions that contribute to growth transformation of primary B cells by EBV. By deleting EBNA1, we have eliminated them all and selected for those attributes essential for growth transformation of infected B cells.
Our study provides genetic evidence that EBNA1 is not an essential transforming gene product of EBV. In vitro, transcriptional activation of potential cellular target genes does not seem to be essential for the establishment of LCLs. All established ΔEBNA1-LCLs proliferate at least for 5 months independently of fibroblast feeder cells indistinguishable of LCLs established with wild-type-EBV (32). Similarly, tumor development in SCID mice injected with LCLs established with EBNA1-expressing or ΔEBNA1-EBV-strains indicated that EBNA1 does not play a role in this model system. These findings are in contrast to a ΔLMP1-EBV-strain that leads to B cell lines, which are not malignant in SCID mice and are dependent on paracrine effects from fibroblast feeder cells for sustained proliferation in vitro (32). Although EBNA1 has been causally associated with different tumors such as Burkitt's lymphomas (among others; ref. 1), our finding is in line with a recent report that failed to establish a role of EBNA1 in a model for this human cancer (48). Multiple reports have shown a correlation between EBNA1 expression and activation of prominent cellular targets such as anti-apoptotic bcl-2 family members, recombinase-activating genes RAG1 and -2, and CD25 (24–26). Tumors observed in EBNA1 transgenic mice have been reported (27, 29) but are controversially discussed.
In light of these reports we cannot exclude that EBNA1 itself has tumorigenic potential, but this study clearly indicates that EBNA1 is not essential for the generation of LCLs by EBV in vitro. EBNA1 enhances the efficiency of this process >10,000-fold by supporting latent viral replication and maintenance of the episomal status of the viral genome. All analyzed ΔEBNA1 cell lines harbor integrated EBV genomes that express at least 5 of the 11 latent EBV genes considered indispensable for this process. The integrity of the viral genome seems to be a critical step, providing a possible explanation for the very low frequency of establishing growth-transformed LCLs with the ΔEBNA1-EBV 2828.
Sporadic reports have always pointed toward EBV-associated tumors in which the viral genome is found chromosomally integrated (39, 49–52). It is remarkable that chromosomal integration of fully functional oncogenic herpesvirus genomes is not uncommon as in the case of T cell lymphomas induced by Marek disease virus (37). It might well be that EBV has yet another mode to transform human cells that is independent of its extrachromosomal state.
Acknowledgments
We thank E. Kremmer for generating and providing the EBNA1, EBNA2, LMP2A, and EBNA3B antibodies and M. Rowe for the EBNA3C-specific antibody. This work has been supported by the Sonderforschungsbereich SFB455 and by additional grants from the Deutsche Forschungsgemeinschaft, by Public Health Service grants CA70723 and CA22443, and by institutional grants.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: EBV, Epstein–Barr virus; LCL, lymphoblastoid cell line; EBNA, EBV nuclear antigen; LMP, latent membrane protein; oriP, origin of replication; SCID, severe combined immunodeficient.
References
- 1.Rickinson, A. B. & Kieff, E. (2001) in Virology, eds. Knipe, D. M. & Howley, P. M. (Lippincott, Philadelphia), Vol. 2, pp. 2575–2627. [Google Scholar]
- 2.Kieff, E. & Rickinson, A. B. (2001) in Virology, eds. Knipe, D. M. & Howley, P. M. (Lippincott, Philadelphia), Vol. 2, pp. 2511–2573. [Google Scholar]
- 3.Leight, E. R. & Sugden, B. (2000) Rev. Med. Virol. 10, 83–100. [DOI] [PubMed] [Google Scholar]
- 4.Aiyar, A., Tyree, C. & Sugden, B. (1998) EMBO J. 17, 6394–6403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sugden, B. (2002) Trends Biochem. Sci. 27, 1–3. [DOI] [PubMed] [Google Scholar]
- 6.Kanda, T., Otter, M. & Wahl, G. M. (2001) Mol. Cell. Biol. 21, 3576–3588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marechal, V., Dehee, A., Chikhi-Brachet, R., Piolot, T., Coppey-Moisan, M. & Nicolas, J. C. (1999) J. Virol. 73, 4385–4392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu, H., Kapoor, P. & Frappier, L. (2002) J. Virol. 76, 2480–2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yates, J. L. (1996) in DNA Replication in Eukaryotic Cells, ed. DePamphilis, M. L. (Cold Spring Harbor Lab. Press, Plainview, NY), pp. 751–773.
- 10.Van Scoy, S., Watakabe, I., Krainer, A. R. & Hearing, J. (2000) Virology 275, 145–157. [DOI] [PubMed] [Google Scholar]
- 11.Wang, Y., Finan, J. E., Middeldorp, J. M. & Hayward, S. D. (1997) Virology 236, 18–29. [DOI] [PubMed] [Google Scholar]
- 12.Shire, K., Ceccarelli, D. F., Avolio-Hunter, T. M. & Frappier, L. (1999) J. Virol. 73, 2587–2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kapoor, P., Shire, K. & Frappier, L. (2001) EMBO J. 20, 222–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fischer, N., Kremmer, E., Lautscham, G., Mueller-Lantzsch, N. & Grasser, F. A. (1997) J. Biol. Chem. 272, 3999–4005. [DOI] [PubMed] [Google Scholar]
- 15.Kim, A. L., Maher, M., Hayman, J. B., Ozer, J., Zerby, D., Yates, J. L. & Lieberman, P. M. (1997) Virology 239, 340–351. [DOI] [PubMed] [Google Scholar]
- 16.Hirai, K. & Shirakata, M. (2001) Curr. Top. Microbiol. Immunol. 258, 13–33. [DOI] [PubMed] [Google Scholar]
- 17.Dhar, S. K., Yoshida, K., Machida, Y., Khaira, P., Chaudhuri, B., Wohlschlegel, J. A., Leffak, M., Yates, J. & Dutta, A. (2001) Cell 106, 287–296. [DOI] [PubMed] [Google Scholar]
- 18.Schepers, A., Ritzi, M., Bousset, K., Kremmer, E., Yates, J. L., Harwood, J., Diffley, J. F. & Hammerschmidt, W. (2001) EMBO J. 20, 4588–4602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang, D., Frappier, L., Gibbs, E., Hurwitz, J. & O'Donnell, M. (1998) Nucleic Acids Res. 26, 631–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Deng, Z., Lezina, L., Chen, C. J., Shtivelband, S., So, W. & Lieberman, P. M. (2002) Mol. Cell 9, 493–503. [DOI] [PubMed] [Google Scholar]
- 21.Hung, S. C., Kang, M. S. & Kieff, E. (2001) Proc. Natl. Acad. Sci. USA 98, 1865–1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gahn, T. A. & Sugden, B. (1995) J. Virol. 69, 2633–2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kang, M. S., Hung, S. C. & Kieff, E. (2001) Proc. Natl. Acad. Sci. USA 98, 15233–15238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Srinivas, S. K. & Sixbey, J. W. (1995) J. Virol. 69, 8155–8158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kube, D., Vockerodt, M., Weber, O., Hell, K., Wolf, J., Haier, B., Grasser, F. A., Muller-Lantzsch, N., Kieff, E., Diehl, V. & Tesch, H. (1999) J. Virol. 73, 1630–1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kuhn-Hallek, I., Sage, D. R., Stein, L., Groelle, H. & Fingeroth, J. D. (1995) Blood 85, 1289–1299. [PubMed] [Google Scholar]
- 27.Wilson, J. B., Bell, J. L. & Levine, A. J. (1996) EMBO J. 15, 3117–3126. [PMC free article] [PubMed] [Google Scholar]
- 28.Wilson, J. B. & Levine, A. J. (1992) Curr. Top. Microbiol. Immunol. 182, 375–384. [DOI] [PubMed] [Google Scholar]
- 29.Tsimbouri, P., Drotar, M. E., Coy, J. L. & Wilson, J. B. (2002) Oncogene 21, 5182–5187. [DOI] [PubMed] [Google Scholar]
- 30.Lee, M. A., Diamond, M. E. & Yates, J. L. (1999) J. Virol. 73, 2974–2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Delecluse, H. J., Pich, D., Hilsendegen, T., Baum, C. & Hammerschmidt, W. (1999) Proc. Natl. Acad. Sci. USA 96, 5188–5193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dirmeier, U., Neuhierl, B., Kilger, E., Reisbach, G., Sandberg, M. L. & Hammerschmidt, W. (2003) Cancer Res. 63, 2982–2989. [PubMed] [Google Scholar]
- 33.Zeidler, R., Meissner, P., Eissner, G., Lazis, S. & Hammerschmidt, W. (1996) Cancer Res. 56, 5610–5614. [PubMed] [Google Scholar]
- 34.Neuhierl, B., Feederle, R., Hammerschmidt, W. & Delecluse, H. J. (2002) Proc. Natl. Acad. Sci. USA 99, 15036–15041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gruffat, H., Batisse, J., Pich, D., Neuhierl, B., Manet, E., Hammerschmidt, W. & Sergeant, A. (2002) J. Virol. 76, 9635–9644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Janz, A., Oezel, M., Kurzeder, C., Mautner, J., Pich, D., Kost, M., Hammerschmidt, W. & Delecluse, H. J. (2000) J. Virol. 74, 10142–10152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Delecluse, H.-J., Schüller, S. & Hammerschmidt, W. (1993) EMBO J. 12, 3277–3286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hammerschmidt, W. & Sugden, B. (1989) Nature 340, 393–397. [DOI] [PubMed] [Google Scholar]
- 39.Delecluse, H. J., Bartnizke, S., Hammerschmidt, W., Bullerdiek, J. & Bornkamm, G. W. (1993) J. Virol. 67, 1292–1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fruehling, S., Lee, S. K., Herrold, R., Frech, B., Laux, G., Kremmer, E., Grasser, F. A. & Longnecker, R. (1996) J. Virol. 70, 6216–6226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Delecluse, H. J., Hilsendegen, T., Pich, D., Zeidler, R. & Hammerschmidt, W. (1998) Proc. Natl. Acad. Sci. USA 95, 8245–8250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kempkes, B., Pich, D., Zeidler, R. & Hammerschmidt, W. (1995) Proc. Natl. Acad. Sci. USA 92, 5875–5879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wendtner, C. M., Kurzeder, C., Theiss, H. D., Kofler, D. M., Baumert, J., Delecluse, H. J., Janz, A., Hammerschmidt, W. & Hallek, M. (2003) Exp. Hematol. 31, 99–108. [DOI] [PubMed] [Google Scholar]
- 44.Farrell, P. J. (2001) Methods Mol. Biol. 174, 3–12. [DOI] [PubMed] [Google Scholar]
- 45.Delecluse, H. J. & Hammerschmidt, W. (1993) J. Virol. 67, 82–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Johannessen, I. & Crawford, D. H. (1999) Rev. Med. Virol. 9, 263–277. [DOI] [PubMed] [Google Scholar]
- 47.Borestrom, C., Zetterberg, H., Liff, K. & Rymo, L. (2003) J. Virol. 77, 821–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kitagawa, N., Goto, M., Kurozumi, K., Maruo, S., Fukayama, M., Naoe, T., Yasukawa, M., Hino, K., Suzuki, T., Todo, S. & Takada, K. (2000) EMBO J. 19, 6742–6750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Henderson, A., Ripley, S., Heller, M. & Kieff, E. (1983) Proc. Natl. Acad. Sci. USA 80, 1987–1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Matsuo, T., Heller, M., Petti, L., O'Shiro, E. & Kieff, E. (1984) Science 226, 1322–1325. [DOI] [PubMed] [Google Scholar]
- 51.Lawrence, J. B., Villnave, C. A. & Singer, R. H. (1988) Cell 52, 51–61. [DOI] [PubMed] [Google Scholar]
- 52.Ohshima, K., Suzumiya, J., Ohga, S., Ohgami, A. & Kikuchi, M. (1997) Int. J. Cancer 71, 943–947. [DOI] [PubMed] [Google Scholar]