Abstract
The rpf gene cluster of Xanthomonas campestris pathovar campestris (Xcc) is required for the pathogenesis of this bacterium to plants. Several rpf genes are involved in the coordinate positive regulation of the production of virulence factors mediated by the small diffusible molecule DSF (for diffusible signal factor). RpfF directs the synthesis of DSF, and a two-component sensory transduction system comprising RpfC and RpfG has been implicated in the perception of the DSF signal and signal transduction. In L medium, rpfF, rpfG, rpfC, and rpfGHC mutants grew as matrix-enclosed aggregates, whereas the wild type grew in a dispersed planktonic fashion. Synthesis of the extracellular polysaccharide xanthan was required for aggregate formation. Addition of DSF triggered dispersion of the aggregates formed by the rpfF strain, but not those of rpf strains defective in DSF signal transduction. An extracellular enzyme from Xcc whose synthesis was positively controlled by the DSF/rpf system could disperse the aggregates produced by all rpf strains. The enzyme was identified as the single endo-β-1,4-mannanase encoded by the Xcc genome. This enzyme had no detectable activity against soluble xanthan. The endo-β-1,4-mannanase was required for the full virulence of Xcc to plants. On the basis of this model system, we propose that one role of the β-mannanase during disease is to promote transitions from an aggregated or biofilm lifestyle to a planktonic lifestyle in response to the DSF signal.
Biofilms have been defined as matrix-enclosed bacterial populations that are adherent to each other and/or to surfaces or interfaces (1). It is evident that many bacterial species live predominantly in biofilms in both natural and artificial environments (1–4). Biofilms are dynamic structures in which transitions between the planktonic and biofilm modes of growth occur presumably in response to different environmental signals. Although the amount of data on the regulation of initiation and maturation of biofilms is increasing, very little is known about the mechanisms of biofilm dispersal or the signals that trigger it (2). Despite the recognition of the ubiquity of biofilms, only relatively few species of bacteria have been studied in any detail. Although most attention has been focused on animal and human pathogens, the ability of plant pathogenic bacteria to form and detach from biofilms may equally have considerable implications for the completion of their disease cycle.
Xanthomonas campestris pathovar campestris (Xcc) is the causal agent of black rot disease of cruciferous plants. The ability of Xcc to incite disease depends on several factors, including the synthesis of extracellular plant cell wall-degrading enzymes and the extracellular polysaccharide (EPS) xanthan (5, 6). The rpf genes act to positively regulate the synthesis of extracellular enzymes, EPS, and pathogenicity (5–8). rpfF, rpfC, and rpfG genes are implicated in a regulatory system involving a diffusible signal factor (DSF). The synthesis of DSF depends on RpfF (7). DSF perception and signal transduction have been suggested to involve the two-component system comprising RpfC and RpfG, which are encoded within the rpfGHC operon (8). RpfH and the products of other genes within the rpf gene cluster (rpfADEI) have no apparent involvement in the DSF regulatory system and have minor regulatory roles.
The work in this paper was prompted by the observation that in certain liquid media, strains with mutations in rpfF, rpfG, and rpfC grow as large multicellular aggregates rather than the dispersed planktonic form seen with the wild-type or other rpf mutants. Examination of the aggregates by scanning electron microscopy revealed that bacteria are held together in a matrix of extracellular material. This knowledge revealed a hitherto unsuspected role for the DSF/rpf regulatory system in biofilm formation and/or dispersal. With this model system, we have identified an enzyme that allows bacteria to escape from an aggregated state in response to the DSF signal and have established a role for this enzyme in the virulence of Xcc to plants.
Materials and Methods
Bacterial Strains and Culture Conditions. Bacterial strains and plasmids and culture conditions and NYGB medium have been described (7–9). L medium contains Bactotryptone (Difco), 10 g·liter–1; yeast extract, 5 g·liter–1; sodium chloride, 5 g·liter–1; and d-glucose, 1 g·liter–1. NYGB medium contains Bacteriological Peptone (Oxoid, Basingstoke, U.K.), 5 g·liter–1; yeast extract (Difco), 3 g·liter–1; and glycerol, 20 g·liter–1.
Enzyme Assays and DSF Preparation. β-1,4-Endoglucanase and protease activities were measured as described (10). β-1,4-Mannanase activity was estimated by radial diffusion assays into substrate-containing agarose plates (11) with locust bean gum (Sigma) as substrate. Degradation of xanthan was estimated both viscometrically and by measurement of the release of reducing sugars. DSF was extracted into ethyl acetate from culture supernatants of Xcc strains grown in NYGB as described (7). The ethyl acetate extracts were evaporated to dryness and resuspended in H2O before use.
Scanning Electron Microscopy. A drop of bacterial suspension was placed on the surface of a piece of moistened cellulose acetate membrane, which had been stuck to the aluminum scanning electron microscope stub by using OCT compound (BDH). Excess liquid was wicked away by using a piece of filter paper but without causing drying. The stub was then immediately plunged into liquid nitrogen slush at approximately –210°C to cryopreserve the material. The sample was transferred onto the cryostage of a CT1500HF cryotransfer system (Gatan, Oxford) attached to a Philips XL30 FEG scanning electron microscope (Philips Electron Optics, Cambridge, U.K.). Sublimation of surface frost was performed at –95°C for 3 min before sputter coating the sample with platinum for 2 min at 10 mA, at colder than –110°C. After sputter coating, the sample was moved onto the cryostage in the main chamber of the microscope, held at approximately –140°C. The sample was viewed at 3 kV, and digital images were saved.
Extracellular Enzymes and Aggregate Dispersal. Extracellular enzyme preparations were obtained from cultures grown in NYGB medium to an OD at 600 nm of 2. Cell-free supernatants were concentrated by addition of ammonium sulfate to a final concentration of 600 g·liter–1. The precipitate that formed at 4°C was harvested by centrifugation and was dialyzed overnight at 4°C against 10 mM Tris·HCl buffer, pH 8. The concentration factor was 20-fold. To observe effects on the dispersal of Xcc aggregate, 5- to 100-μl aliquots of the dialyzed preparations were added to 5 ml of 50-ml overnight cultures of rpf mutants grown in L broth dispensed into universal bottles and maintained at 30°C. The percentage of the bacteria in the culture that were present in aggregates was estimated as follows. After 5 min without shaking, when the aggregated bacteria had settled, a 1-ml sample was removed from the top of the culture and the number of (planktonic) bacteria was estimated by measurement of OD at 600 nm. To estimate the total bacteria in the culture, a preparation of the dispersing enzyme was added. After 20 min at 30°C, with periodic vortexing, all the aggregates had dispersed and the total number of bacteria in the culture could be estimated by measurement of OD at 600 nm of an appropriate dilution.
Purification of Enzyme Activity Capable of Dispersing Aggregates. The enzyme activity capable of dispersing aggregates was partially purified from culture supernatants of the engXCA strain 8409 (10). This strain was chosen as a source of the enzyme to eliminate the presence of the endoglucanase EngXCA, which constitutes >90% of the total extracellular protein in the wild type (10), but which has no activity against aggregates. This choice was to allow easier analysis of less abundant proteins. Cell-free culture supernatants were treated with 2 mM EDTA, 2 mM PMSF, and 1 mM 1,10-phenathroline for 2 h at 25°C before concentration with ammonium sulfate and dialysis. After dialysis the sample was passed through a column of DEAE-Sephadex equilibrated with 20 mM Tris·HCl buffer, pH 8, and eluted with the same buffer. The eluate (which contained all of the activity) was dialyzed overnight against distilled water, and 1 M sodium acetate buffer, pH 5, was added to give a final concentration of 20 mM. This sample was fractionated by ion-exchange perfusion chromatography on a CM-cellulose column by using the BioCad Sprint apparatus (PerSeptive Biosystems, Framingham, MA). The column was equilibrated with 20 mM sodium acetate buffer, pH 5, and all the activity bound to the column at this pH. The column was then eluted with a linear gradient of 0–1 M NaCl in 20 mM sodium acetate buffer, pH 5, in a total volume of 40 ml and at a flow rate of 2ml·min–1. One-milliliter fractions were collected and assayed for the ability to disperse aggregates.
Peptide Mass Fingerprinting of Proteins. Bands were cut from the SDS-polyacrylamide gel stained with Coomassie blue, and the proteins were reduced, derivatized with iodoacetamide, and subjected to enzymatic hydrolysis with trypsin. The sizes of the peptide fragments generated were determined by mass spectrometry on a Bruker Reflex III matrix-assisted laser desorption ionization–time-of-flight mass spectrometer (Bruker Daltonics, Coventry, U.K.). The data were used to interrogate the National Center for Biotechnology Information nonredundant protein databases by using the mascot program available at www.matrixscience.com. For band 1, the only significant hit (probability-based mowse score of 94) was a pectate lyase from Xcc (GenBank accession no. AE012162), where eight peptides matched. For band 2, the only significant hit (probability-based mowse score of 85) was mannan endo-β-1,4-mannosidase from Xcc (GenBank accession no. AE012279), where eight peptides matched. Protein scores >72 indicate that the observed match is significant, where the probability (P) of a random match is <0.05.
Creation of Mutant in the Gene Encoding Mannan Endo-β-1,4-mannosidase. Strains in which the manA gene encoding the endo-β-1,4-mannanase was disrupted were created with the use of the plasmid pK18mob (12). A 491-bp internal fragment of the manA gene was amplified by PCR using Xcc chromosomal DNA as template and the primers ManF2, 5′-GCTGGAACCGCACGCCCGAGG-3′, and ManR2, 5′-ACCAGCGCAAGATTGTTGCTG-3′, and cloned by using the TOPO TA cloning kit (Invitrogen). The identity of the cloned fragment was confirmed by sequencing. The fragment was excised from this construct by using EcoRI, which cuts at sites flanking the insert. This EcoRI fragment was ligated into pK18mob to create pK18manS. This construct was conjugated from Escherichia coli DH5α into Xcc by triparental mating by using the helper plasmid pRK2073. Mutants were selected on plates containing rifampicin (50 μg/ml) and kanamycin (12.5 μg/ml). Mutants were screened for disruption of the manA gene by PCR and by Southern blotting. manA is flanked by genes that are transcribed in the opposite orientation; hence, disruption of manA does not have polar effects.
Virulence Testing. The virulence of Xcc to Chinese radish (Raphanus sativus variety niger) was estimated after bacteria were introduced into the leaves by infiltration, leaf clipping, or spraying. Bacteria grown overnight in NYGB medium were washed and resuspended in water to an OD at 600 nm of 0.1, 0.001, or 0.0001. For leaf clipping the last completely expanded leaf was cut with scissors dipped in bacterial suspensions of OD 0.1 or 0.001. Lesion length was measured 14 days after inoculation on 15 leaves for each strain tested. For spraying, bacterial suspensions of OD 0.1 or 0.001 were applied to between 40 and 60 plants per treatment. Lesions appeared at the leaf margin, and the number of plants with lesions was measured 14 days after inoculation. For infiltration inoculation bacterial suspensions of OD of 0.1, 0.001, or 0.0001 were introduced into the leaves by using a syringe.
Results
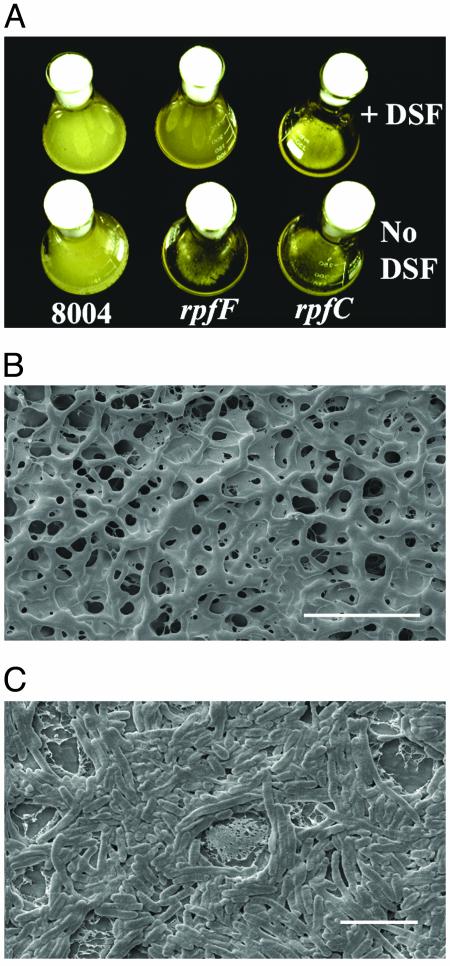
Formation of Cell Aggregates by rpf Mutants of X. campestris. Strains of Xcc carrying mutations in genes within the rpf gene cluster grow in a dispersed fashion in NYGB medium with growth characteristics similar to the wild type. However in L medium, rpfF, rpfG, and rpfC mutants and the triple deletion mutant rpfGHC grew in an aggregated state, forming readily observable clumps (Fig. 1A). The wild-type and other rpf mutants (rpfA, rpfB, rpfE, rpfH, and rpfI) did not form such aggregates in L medium and grew in a dispersed fashion. After 24 h of growth in L medium, >95% of the bacteria in the cultures of rpfF, rpfG, rpfC, and rpfGHC mutants were present in aggregates, although with extended culture times, an increasing proportion of planktonic bacteria were seen. Examination by scanning electron microscopy indicated that bacteria within aggregates were held together in a matrix of extracellular material to form a three-dimensional reticulated structure (Fig. 1B). In contrast, bacteria growing in a dispersed fashion showed no aggregation by light microscopy and were not embedded in an extracellular matrix (Fig. 1C). Supplementation of L medium with DSF before bacterial inoculation allowed for dispersed growth of the rpfF mutant (Fig. 1 A). The growth characteristics of rpfG, rpfC, and rpfGHC mutant strains were not affected by addition of DSF, however (Fig. 1 A). Addition of DSF to 24-h cultures of the rpfF mutant in L medium triggered the dispersal of the aggregates, so that within3hofDSF addition, almost all the bacteria within the culture were in the planktonic state. Under the same conditions, rpfG, rpfC, and rpfGHC strains remained in the aggregated state. This finding established that dispersal is triggered by DSF, acting through the RpfC/RpfG two-component signaling system.
Fig. 1.
Formation of aggregates by X. campestris and the role of DSF. (A) Strains of X. campestris with mutations in rpfF, rpfG, and rpfC grow in an aggregated fashion in L medium, whereas the wild-type strain (8004) grows in a dispersed fashion. Addition of the DSF to rpfF strains but not to rpfG or rpfC strains causes dispersed growth. (B) Scanning electron microscopy reveals that aggregates of rpf strains have a reticulated structure where bacteria are held within a matrix. (Scale bar = 10 μm.) (C) Wild-type bacteria in contrast show no such matrix encasement. (Scale bar = 5 μm.) Note that these bacteria showed no aggregation in light microscopy before application to the membrane used in scanning electron microscopy.
Formation of Aggregates Requires the EPS Xanthan. An important role for EPS in the formation of biofilms has already been established for several bacteria (13–17). To examine the role of xanthan in aggregate formation, we created strains that could not produce the polysaccharide in an rpf mutant background. The gum gene cluster composed of 12 genes, gumB to gumM, directs the synthesis and export of xanthan (18, 19). A transposon insertion located 15 bp upstream of the translational start site of gumB almost completely eliminates the production of xanthan (20). Introduction of this Tn5 insertion into the chromosome of rpfG and rpfGHC mutants generated the mutant strains rpfG gum and rpfGHC gum. As expected, these strains did not produce detectable xanthan in NYGB medium supplemented with 2% glucose. Under the same conditions the rpf mutant strains produce between 30% and 50% of the wild-type level of xanthan (20). Significantly, the rpfG gum and rpfGHC gum strains were unable to form aggregates in L medium, and no EPS was recovered by ethanol precipitation from culture supernatants in L medium. These results indicated that the synthesis of xanthan is necessary for the formation of aggregates by rpf mutants of Xcc.
Extracellular Enzyme Activity of Xcc Capable of Dispersing Aggregates. Aggregates of bacteria could not be dispersed by resuspension and vortexing in salt solutions (up to 0.2 M KCl), or in 1 mM EDTA. Addition of proteinase K (50 μg/ml) or lysozyme (50 μg/ml) to the cultures for 6 h also had no effect. Aggregates could be dispersed without loss of bacterial viability by a preparation of extracellular proteins from the wild-type Xcc (Fig. 2 A and B). The activity was destroyed by treatment at 100°C for 5 min, suggesting that it was enzymatic in nature. Examination of the dispersed cultures by light microscopy and scanning electron microscopy indicated that the bacteria were intact. The bacteria were also viable as determined by plating. Extracellular enzyme preparations from rpf mutants had, by contrast, very little activity against the aggregates. Concentrated supernatants from cultures of the rpfF mutant supplemented with DSF had almost wild-type levels of the dispersing enzyme activity. These results established that the synthesis of the enzyme(s) promoting dispersal was under control of the rpf genes and DSF. The dispersing activity was insensitive to inhibition by 1 mM PMSF, 5 mM EDTA, 2 mM iodoacetamide, 14 μM pepstatin, or 2 mM 1,10-phenanthroline. Treatment with these protease inhibitors either alone or in combination for1hat25°C did not lead to loss of activity. Supernatants from Xcc mutants with transposon insertions in engXCA, encoding the major extracellular β-1,4-endoglucanase (10), or egl2, encoding a minor β-1,4-endoglucanase (10), were equally effective as the wild type in dispersion of aggregates. We concluded that the dispersing activity was not due to either of the two predominant extracellular β-1,4-endoglucanases acting alone.
Fig. 2.
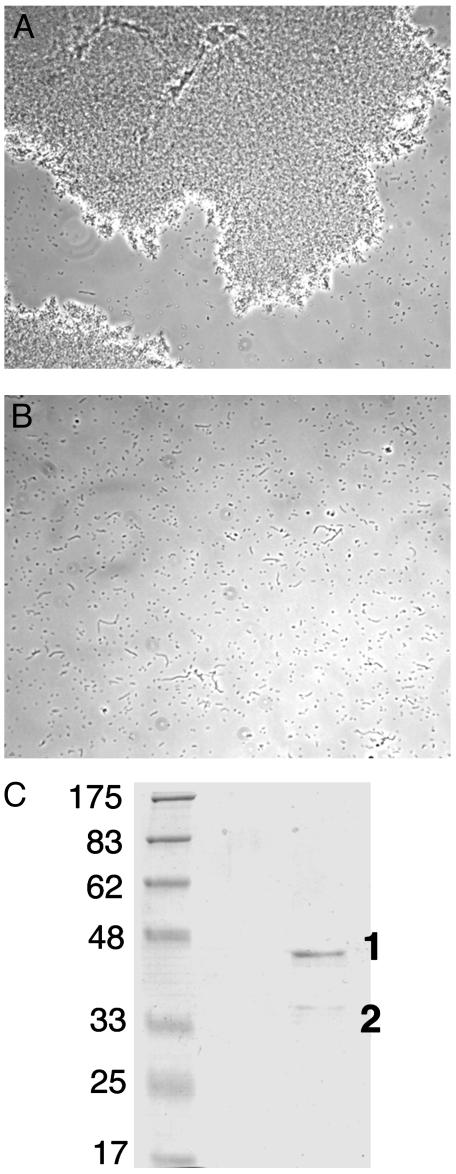
An extracellular enzyme from X. campestris can disperse the aggregates. (A) Light microscopy of cultures of an rpf mutant after 24 h of growth. (B) The same culture after treatment for 30 min with the dispersing enzyme. (C) Coomassie blue-stained SDS/PAGE of the most active fraction from ion-exchange chromatography of the dispersing enzyme preparation. Molecular mass standards are given in kilodaltons; protein bands 1and 2 are indicated.
Characterization of Enzyme Activity. The enzyme activity was partially purified from culture supernatants by ion-exchange chromatography. On CM-cellulose cation-exchange chromatography at pH 5, a peak of activity was seen that eluted from the column at ≈0.5 M NaCl and was resolved from a β-1,4-endoglucanase activity that eluted at 0.3 M NaCl. On SDS/PAGE, all active fractions had a major band (band 1) with an apparent molecular mass of 44 kDa and a minor band (band 2) with an apparent molecular mass of 35 kDa (Fig. 2C). Addition of this preparation to cultures to give a final concentration of 750 ng of protein per ml caused dispersal of aggregates within 20 min.
The proteins in bands 1 and 2 were identified by peptide mass fingerprinting as a pectate lyase (AE012162) and the single mannan endo-β-1,4-mannosidase (AE012279) of Xcc, respectively (see Materials and Methods). Both predicted proteins have cleavable signal sequences, consistent with their extracellular location. The predicted molecular mass of the endo-β-1,4-mannosidase after signal sequence cleavage is 33,129 Da, which is close to the value estimated by SDS/PAGE. For the pectate lyase, the predicted mass of the protein after signal sequence cleavage was 37,620 Da. The discrepancy with the mass estimated from SDS/PAGE could arise because of anomalous electrophoretic behavior of this protein.
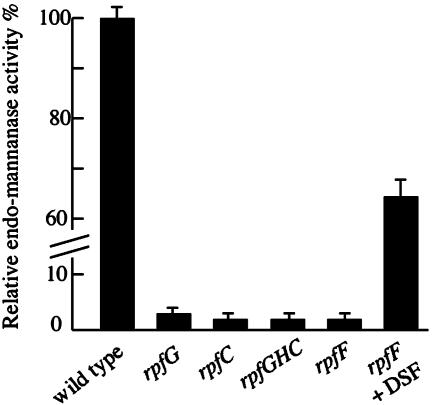
To establish which of the two candidate enzymes was responsible for dispersal, we studied strains carrying mutations in the cognate genes. Two strains carrying different Tn5gus insertions within the gene for the pectate lyase were identified within a collection of such mutants generated in one of our laboratories. Concentrated culture supernatants from these strains were equally effective as the wild type in aggregate dispersal. Strains deficient in endo-β-1,4-mannanase were generated by disruption of the cognate gene (manA) by the use of pK18mob carrying an internal fragment of the manA gene (see Materials and Methods). Concentrated culture supernatants from the manA strain were not able to disperse the aggregates. The manA mutant had lost endo-β-1,4-mannanase activity (as expected) although the levels of other extracellular enzymes (β-1,4-endoglucanase and protease) were unaffected. Culture supernatants from rpf mutants in NYGB medium had relatively little endo-β-1,4-mannanase activity, although addition of DSF to cultures of the rpfF mutant partially restored production (Fig. 3). In L medium the wild type produced only 5% of the level of endo-β-1,4-mannanase seen in NYGB medium, and activity was not detectable in culture supernatants of rpf mutants. Addition of DSF to the rpfF mutant partially restored production, however (data not shown). Overall these results establish that the dispersing enzyme is the single endo-β-1,4-mannanase of Xcc and that the synthesis of this enzyme is controlled by the DSF/rpf system.
Fig. 3.
Synthesis of the endo-β-1,4-mannanase is positively regulated by the rpf genes in NYGB medium. Addition of DSF restores enzyme production to the rpfF mutant. In L medium, enzyme activity was not detected in culture supernatants of rpf mutants.
The ability of the crude concentrated supernatant and partially purified enzyme to degrade mature xanthan and deacetylated xanthan was estimated by viscometric measurements and by measurements of release of reducing sugars. Both types of assay gave no indication of degradation of either substrate.
Endo-β-1,4-mannanase Contributes to Bacterial Virulence. Wild-type and manA strains of Xcc were applied to leaves of Chinese radish by leaf clipping, spraying, or direct infiltration into leaf panels. Xcc is a vascular pathogen and is normally restricted to the xylem of leaves of infected plants at the early stages of disease. Bacteria enter unwounded plants through hydathodes at the leaf margin. After spraying, V-shaped lesions appeared at the leaf margins at vein endings, indicating that bacteria have entered via hydathodes. Leaf clipping introduces the bacteria directly into the vascular system, whereas infiltration introduces the bacteria into the mesophyllic tissue of the leaf.
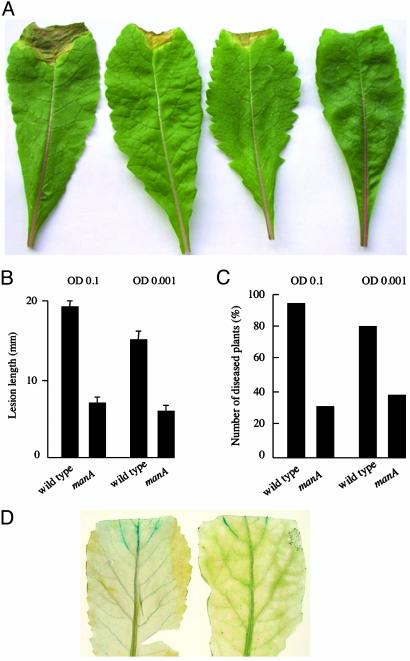
The manA mutant showed markedly reduced virulence compared with the wild type when the bacteria were introduced into the vascular system by leaf clipping or entered the vascular system after spraying. The reduced virulence was indicated by a reduced number of infected plants after spraying and a shorter lesion length than the wild type after both spraying and leaf clipping (Fig. 4). In contrast, when the bacteria were inoculated into the leaf mesophyll tissue, no differences in symptom production between the manA mutant and wild type were observed (data not shown). Comparisons of manA and wild-type strains constitutively expressing β-glucuronidase showed that the manA mutant was impaired in spread along the vascular system (Fig. 4). Taken together these findings indicate that the endo-β-1,4-mannanase contributes to black rot disease progression.
Fig. 4.
The endo-β-1,4-mannanase is required for full virulence of X. campestris to plants. (A) Symptom production on leaves 14 days after inoculation by clipping with (from the left) wild type, two manA mutant strains (derived independently), and water (mock inoculation). (B) The average lesion lengths caused by the manA mutant are significantly shorter (P < 0.01) than those by the wild type at two different inoculation levels (indicated by OD at 600 nm of 0.1 and 0.001). Values are the mean ± SD from 15 measurements. (C) The percentage of diseased plants is also lower after spray inoculation with the manA mutant than with the wild type at two inoculum levels. Results from a typical experiment are shown. This experiment and that shown in B were repeated two more times with essentially the same result. (D) The manA mutant (right leaf) shows reduced spread through the vascular system of infected plants compared with the wild type (left leaf) as demonstrated by staining for bacteria constitutively expressing β-glucuronidase. Infected leaves were stained for β-glucuronidase activity at 4 days after leaf clipping.
Discussion
Cell–Cell Signaling and Biofilm Formation and Dispersal. Cell–cell signaling mediated by diffusible molecules is known to play a large role in regulating diverse physiological processes, including the formation and dispersal of biofilms, across distant genera of bacteria. Quorum sensing mediated by N-acylhomoserine lactones (N-AHLs) has been implicated in the regulation of biofilm formation or aggregation in a number of bacteria (21–26), although it appears to have divergent regulatory roles in different bacteria. In Pseudomonas aeruginosa, N-(3-oxododecanoyl)-homoserine lactone (HSL) is required for the development of mature biofilms from the microcolonies that form after bacterial cell attachment and proliferation on a surface (21). Similarly, in Burkholderia cepacia, N-octanoyl-HSL is implicated in biofilm maturation (22). In Pseudomonas fluorescens addition of N-hexanoyl-HSL accelerates and increases biofilm formation (23). In contrast to the promotion of biofilm formation in these bacteria, quorum sensing in Yersinia pseudotuberculosis, Rhodobacter sphaeroides, and Pseudomonas aureofaciens has been implicated in the maintenance of the dispersed state. Disruption of a hierarchical quorum-sensing system in Y. pseudotuberculosis by mutation of ypsR leads to the formation of clumps in liquid media (24). In R. sphaeroides, mutation of cerI, which directs synthesis of N-(7,8-cis-tetradecenoyl)-HSL, leads to the formation of large aggregates of bacteria in liquid culture (25). This aggregated phenotype in R. sphaeroides can be reversed by addition of the N-AHL signaling molecule. Similar effects are seen in P. aureofaciens after mutation of csaI and after addition of the cognate N-AHL (26). Although DSF is not an N-AHL, the regulatory role of the DSF system on aggregate dispersal in Xcc has superficial parallels with N-AHL-mediated regulation of aggregate dispersal in Y. pseudotuberculosis, P. aureofaciens, and R. sphaeroides. However, the processes that are regulated and that promote aggregate dispersal in these other bacteria have not yet been defined and are probably different in each case. For example, aggregation by cerI mutants of R. sphaeroides is believed to occur as a consequence of overproduction of EPS, whereas in X. campestris, mutation of rpfF and consequent loss of the synthesis of DSF has the effect of reducing xanthan production.
EPS and Biofilms. An important role for EPS in the formation of biofilms has already been established for several bacteria (13–17). Here we have provided genetic evidence that the synthesis of the extracellular polysaccharide xanthan is important for the integrity of Xcc aggregates in culture. In NYGB cultures of Xcc, xanthan is synthesized and released as a soluble polysaccharide into the growth medium. The mechanism by which xanthan is retained and contributes to structural integrity of the aggregates of rpf mutants in L medium is unknown. As discussed by Sutherland (3), potential mechanisms could include chain–chain entanglements, interchain helix formation, interaction of the polysaccharide molecules with surface-attached components such as proteins or glycoproteins or interactions with other polysaccharide molecules. The characterization of the Xcc dispersing enzyme as an endo-β-(1,4)-mannanase is particularly intriguing in the context of xanthan interaction with other polysaccharides, because it has been long established that xanthan forms gels when mixed with polymers possessing a β-(1,4)-mannan backbone (27) and molecular interactions between the two types of polymer have been studied extensively in vitro (28). The precise substrate for the endo-β-(1,4)-mannanase is not known however. There are no reports of the synthesis of β-1,4-mannans by Xcc that could be a biofilm-specific process that is negatively regulated by the DSF/rpf system. The manA mutant grows in a dispersed fashion in L medium, suggesting that down-regulation of the endo-β-(1,4)-mannanase activity is necessary but not sufficient for the formation of aggregates.
Enzymes and Biofilm Dispersal. It has been shown previously that the release of bacteria from biofilms can involve enzymatic degradation of the EPS component. In P. aeruginosa, degradation of alginate by the alginate lyase of this bacterium contributes to cell detachment from surfaces (29). Enzymatic degradation of a biofilm-specific EPS component has been implicated in detachment of P. fluorescens from surfaces (23), although the nature of the enzyme and its substrate and the mode of regulation of this process are unclear. Similarly, the dispersal of Xcc aggregates does not require degradation of xanthan, the major EPS of planktonic cultures, but of a second component that may be a biofilm-specific EPS. Only one gene for endo-β-1,4-mannosidase exists in the chromosome of Xcc (ref. 30; J.-L.T., personal communication), and an almost identical gene product is found in Xanthomonas axonopodis pathovar citri (30).
Biofilms and Black Rot Disease. Mutants of Xcc with defects in xanthan production are known to have severely reduced virulence in plants (31–33). Our in vitro experiments suggest that one possible role of xanthan in the disease process is in the formation of biofilms, which might promote bacterial survival against stresses such as desiccation and the action of plant-derived antimicrobial compounds. Biofilm formation in vitro has been demonstrated for Xylella fastidiosa (34), a pathogen related to Xcc that in nature colonizes only two niches; the xylem vessels of the plant host and the feeding mouthparts and foregut of its insect vector. It has similarly been proposed that X. fastidiosa bacteria are aggregated in biofilms in these locations (34).
Our in planta studies established that the endo-β-1,4-mannanase is required for the full virulence of Xcc to Chinese radish. On the basis of our in vitro studies, we suggest that one role of the enzyme in disease is to allow the release of bacteria from an aggregated or biofilm lifestyle to a planktonic lifestyle to promote the colonization of the vascular system. We propose that DSF acts as an environmental signal in planta to trigger such transitions in bacterial lifestyle. We cannot, however, exclude a second role for the enzyme, to mobilize mannan hemicelluloses of the plant cell wall for nutritional purposes.
The DSF/rpf regulatory system appears to be restricted to Xanthomonas and the closely related genera Xylella (6) and Stenotrophomonas (D. Murphy and J.M.D., unpublished data). It remains to be seen whether this system regulates processes related to biofilm dispersal in these other bacteria.
Acknowledgments
The Sainsbury Laboratory is supported by a grant from the Gatsby Charitable Foundation. The work was also supported by the Biotechnology and Biological Sciences Research Council (Grant PRS12153) and the Science Foundation of Ireland, through the award of an E. T. S. Walton Fellowship (to J.M.D.). The work was carried out according to the provisions of the Ministry of Agriculture, Fisheries and Food License PHF 1185/8 (48) issued under the Plant Health (Great Britain) Order 1987 (Statutory Instrument 1758).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: Xcc, Xanthomonas campestris pathovar campestris; EPS, extracellular polysaccharide; DSF, diffusible signal factor.
References
- 1.Costerton, J. W., Lewandowski, Z., Caldwell, D. E., Korber, D. R. & Lappin-Scott, H. M. (1995) Annu. Rev. Microbiol. 49, 711–745. [DOI] [PubMed] [Google Scholar]
- 2.O'Toole, G., Kaplan, H. B. & Kolter, R. (2000) Annu. Rev. Microbiol. 54, 49–79. [DOI] [PubMed] [Google Scholar]
- 3.Sutherland, I. W. (2001) Microbiology 147, 3–9. [DOI] [PubMed] [Google Scholar]
- 4.Sutherland, I. W. (2001) Trends Microbiol. 9, 222–227. [DOI] [PubMed] [Google Scholar]
- 5.Tang, J. L., Liu, Y. N., Barber, C. E., Dow, J. M., Wootton, J. C. & Daniels, M. J. (1991) Mol. Gen. Genet. 226, 409–417. [DOI] [PubMed] [Google Scholar]
- 6.Dow, J. M. & Daniels, M. J. (2000) Yeast 17, 263–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barber, C. E., Tang, J. L., Feng, J. X., Pan, M. Q., Wilson, T. J. G., Slater, H., Dow, J. M., Williams, P. & Daniels, M. J. (1997) Mol. Microbiol. 24, 555–566. [DOI] [PubMed] [Google Scholar]
- 8.Slater, H., Alvarez-Morales, A., Barber, C. E., Daniels, M. J. & Dow, J. M. (2000) Mol. Microbiol. 38, 986–1003. [DOI] [PubMed] [Google Scholar]
- 9.Turner, P., Barber, C. & Daniels, M. (1985) Mol. Gen. Genet. 199, 338–343. [Google Scholar]
- 10.Gough, C. L., Dow, J. M., Barber, C. E. & Daniels, M. J. (1988) Mol. Plant–Microbe Interact. 1, 275–281. [DOI] [PubMed] [Google Scholar]
- 11.Bourgault, R. & Bewley, J. D. (2002) Anal. Biochem. 300, 87–93. [DOI] [PubMed] [Google Scholar]
- 12.Schafer, A., Tauch, A., Jager, W., Kalinowski, J., Thierbach, G. & Pühler, A. (1994) Gene 145, 69–73. [DOI] [PubMed] [Google Scholar]
- 13.Boyd, A. & Chakrabarty, A. M. (1995) J. Ind. Microbiol. 15, 162–168. [DOI] [PubMed] [Google Scholar]
- 14.Hellman, C., Schweitzer, O., Gerke, C., Vanittanakom, N., Mack, D. & Götz, F. (1996) Mol. Microbiol. 20, 1083–1091. [DOI] [PubMed] [Google Scholar]
- 15.Yilditz, F. H. & Schoolnik, G. K. (1999) Proc. Natl. Acad. Sci. USA 96, 4028–4033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Danese, P. N., Pratt, L. A. & Kolter, R. (2000) J. Bacteriol. 182, 3593–3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zogaj, X., Nimtz, M., Rohde, M., Bokranz, W. & Röhmling, U. (2001) Mol. Microbiol. 39, 1452–1463. [DOI] [PubMed] [Google Scholar]
- 18.Harding, N. E., Cleary, J. M. & Ielpi, I. (1995) in Food Biotechnology: Microorganisms, eds. Hui, Y. H. & Khachatourians, G. G. (Am. Soc. Microbiol., Washington, DC), pp. 253–262.
- 19.Vanderslice, R. W., Doherty, D. H., Capage, M. A., Betlach, M. R., Hassler, R. A., Henderson, N. M., Ryan-Graniero, J. & Tecklenburg, M. (1990) in Biomedical and Biotechnological Advances in Industrial Polysaccharides, eds. Crescenzi, V., Dea, I. C. M., Paoletti, S., Stivala, S. S. & Sutherland, I. W. (Gordon and Breach, New York), pp. 145–156.
- 20.Vojnov, A. A., Zorreguieta, A., Dow, J. M., Daniels, M. J. & Dankert, M. A. (1998) Microbiology 144, 1487–1493. [DOI] [PubMed] [Google Scholar]
- 21.Davies, D. G., Parsek, M. R., Pearson, J. P., Iglewski, B. H. & Greenberg, E. P. (1998) Science 280, 295–298. [DOI] [PubMed] [Google Scholar]
- 22.Huber, B., Riedel, K., Hentzer, M., Heydorn, A., Gotslich, A., Givskov, M., Molin, S. & Eberl, L. (2001) Microbiology 147, 2517–2528. [DOI] [PubMed] [Google Scholar]
- 23.Allison, D. G., Ruiz, B., San Jose, C., Jasope, A. & Gilbert, P. (1998) FEMS Microbiol. Lett. 167, 179–184. [DOI] [PubMed] [Google Scholar]
- 24.Atkinson, S., Throup, J. P., Stewart, G. S. & Williams, P. (1999) Mol. Microbiol. 33, 1267–1277. [DOI] [PubMed] [Google Scholar]
- 25.Puskas, A., Greenberg, E. P., Kaplan, S. & Schaefer, A. L. (1997) J. Bacteriol. 179, 7530–7537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang, Z. & Pierson, L. S., III (2001) Appl. Environ. Microbiol. 67, 4305–4315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morris, E. R., Rees, D. A., Young, G., Walkinshaw, M. D. & Darke, A. (1977) J. Mol. Biol. 110, 1–16. [DOI] [PubMed] [Google Scholar]
- 28.Bresolin, T. M., Milas, M., Rinaudo, M., Reicher, F. & Ganter, J. L. (1999) Int. J. Biol. Macromol. 26, 225–231. [DOI] [PubMed] [Google Scholar]
- 29.Boyd, A. & Chakrabarty, A. M. (1994) Appl. Environ. Microbiol. 60, 2355–2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.da Silva, A. C., Ferro, J. A., Reinach, F. C., Farah, C. S., Furlan, L. R., Quaggio, R. B., Monteiro-Vitorello, C. B., Van Sluys, M. A., Almeida, N. F., Alves, L. M., et al. (2002) Nature 417, 459–463. [DOI] [PubMed] [Google Scholar]
- 31.Newman, M.-A., Conrads-Strauch, J., Scofield, G., Daniels, M. J. & Dow, J. M. (1994) Mol. Plant–Microbe Interact. 5, 553–563. [DOI] [PubMed] [Google Scholar]
- 32.Chou, F.-L., Chou, H.-C., Lin, Y.-S., Yang, B.-Y., Lin, N.-T., Weng, S.-F. & Tseng, Y.-H. (1997) Biochem. Biophys. Res. Commun. 233, 265–269. [DOI] [PubMed] [Google Scholar]
- 33.Katzen, F., Ferreiro, D. U., Oddo, C. G., Ielmini, M. V., Becker, A., Puhler, A. & Ielpi, L. (1998) J. Bacteriol. 180, 1607–1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marques, L. L. R., Ceri, H., Manfio, G. P., Reid, D. M. & Olsen, M. E. (2002) Plant Dis. 86, 633–638. [DOI] [PubMed] [Google Scholar]