Abstract
BMS-378806 is a recently discovered small molecule HIV-1 inhibitor that blocks viral entrance to cells. The compound exhibits potent inhibitory activity against a panel of R5-(virus using the CCR5 coreceptor), X4-(virus using the CXCR4 coreceptor), and R5/X4 HIV-1 laboratory and clinical isolates of the B subtype (median EC50 of 0.04 μM) in culture assays. BMS-378806 is selective for HIV-1 and inactive against HIV-2, SIV and a panel of other viruses, and exhibits no significant cytotoxicity in the 14 cell types tested (concentration for 50% reduction of cell growth, >225 μM). Mechanism of action studies demonstrated that BMS-378806 binds to gp120 and inhibits the interactions of the HIV-1 envelope protein to cellular CD4 receptors. Further confirmation that BMS-378806 targets the envelope in infected cells was obtained through the isolation of resistant variants and the mapping of resistance substitutions to the HIV-1 envelope. In particular, two substitutions, M426L and M475I, are situated in the CD4 binding pocket of gp120. Recombinant HIV-1 carrying these two substitutions demonstrated significantly reduced susceptibility to compound inhibition. BMS-378806 displays many favorable pharmacological traits, such as low protein binding, minimal human serum effect on anti-HIV-1 potency, good oral bioavailability in animal species, and a clean safety profile in initial animal toxicology studies. Together, the data show that BMS-378806 is a representative of a new class of HIV inhibitors that has the potential to become a valued addition to our current armamentarium of antiretroviral drugs.
With the exception of the recently approved fusion inhibitor enfuvirtide (T-20), all of the currently approved HIV drugs target two viral enzymes: reverse transcriptase and protease (1). Despite the significant success achieved with antiretroviral combination therapies, the emergence of resistant viruses and lack of patient compliance stemming from adverse side effects and complex regimens have resulted in many therapeutic failures (2). In addition, close to 20% of newly diagnosed HIV patients are infected with viruses resistant to existing drug classes (3, 4). Therefore, the availability of better tolerated antiretroviral agents that function through novel mechanisms and lack cross-resistance to the existing drugs will be essential to the future management of HIV infection.
The viral entry process provides new anti-HIV-1 targets, with potential for novel classes of drugs (5, 6). The HIV envelope consists of an exterior glycoprotein gp120 and a transmembrane domain gp41, both of which are processed from the gp160 precursor (7, 8). Sequence comparisons revealed that the gp120 glycoprotein is defined by five variable regions (V1 to V5) interspersed with five conserved regions (C1 to C5) (9). Because of the genetic heterogeneity exhibited by HIV-1 envelope proteins, clinical isolates are classified into three distinct groups, M for main, O for outlier, and N for new or non-M (10, 11). The majority of AIDS patients are infected with HIV-1 strains of the M family. Group M can be further divided into subtypes A to K based on genetic similarities (10, 11). Significant efforts on vaccine development have resulted in a better understanding on the geographical distribution of the viral subtypes (12).
HIV entry involves the initial attachment of gp120 with cellular CD4 receptors. Subsequent conformational changes facilitate the binding of gp120 to a chemokine coreceptor, either CCR5 or CXCR4, followed by additional structural adjustments that enable the insertion of the fusion peptide into the host membrane and finally, fusion of the membranes by a still unresolved mechanism (7, 8). Structural information of the HIV-1 envelope has been provided by x-ray crystallography studies of the gp120 core protein complexed with a two-domain fragment of CD4 and a monoclonal antibody (13, 14). Proof of concept for entry targets has been obtained in clinical trials using the attachment inhibitor PRO542 (a CD4-IgG fusion protein) (15, 16), the CCR5 coreceptor antagonist SCH-C,¶ and the fusion inhibitor enfuvirtide (17–19). In this study, a small molecule inhibitor of viral entry, BMS-378806, is characterized with regards to its antiviral activity, inhibition mechanism, resistance profile, and preliminary pharmacokinetic properties.
Materials and Methods
Compound. BMS-378806 was synthesized at Bristol-Myers Squibb. [3H]BMS-378806 was prepared at VITrax (Placentia, CA) and had a specific activity of 38 Ci/mmol (1 Ci = 37 GBq).
Virus and Cells. Cell lines (MT-2, MT-4, PM1, HeLa-CD4, and U87-CD4), HIV-1 laboratory strains (LAI, HXB2, JRFL, Ba-L, MN, IIIB, AO-18, SF-2, and SF-162), and the majority of clinical isolates were from the National Institutes of Health AIDS Reagent Repository. HIV-189.6 was from Advanced Biotechnologies (Columbia, MD). HepG2, human foreskin fibroblast, U373-MG, SK-N-MC, SK-N-SH, HT-29, 293, and Huh-7 cells were from the American Type Culture Collection.
Cytotoxicity. Cytotoxicity assays were performed in the presence of serially diluted inhibitors for 6 days, and cell viability was quantitated by using an XTT assay (20) to assess HepG2, Huh-7, 293, human foreskin fibroblast, HeLa-CD4, U373-MG, SK-N-MC, SK-N-SH, HT-29, PM1, MT-2, MT-4, macrophages, and peripheral blood mononuclear cells (PBMCs).
Time-of-Addition Study. Experiments were done by using a single-cycle viral infection assay (21). HIV-1 with a luciferase reporter gene and pseudotyped with an HIV-1JRFL envelope was used to infect HeLa CD4+ CCR5+ cells in the presence of 50 nM BMS-378806 or 1 μM nevirapine (10 times their respective EC50 values). The compounds were applied at various times after infection. Antiviral activity was determined by quantitating luciferase activity (Promega).
Chemokine Coreceptor Studies. U87-CD4 cells (22) were transfected with an expression vector encoding CCR5 or CCR3. The next day, cells were plated in the presence of increasing concentrations of BMS-378806 and infected with an HIV-1 ADA-Renilla luciferase reporter virus (23). The luciferase activity was measured after 3 days of incubation.
gp120/CD4 ELISA Binding Assay. Assay plates were coated with 1 μg per well of D7324 (Aalto Bio Reagents, Dublin), a sheep antibody to the C terminus of HIV-1LAI gp120 (24). Coated plates were applied with gp120 at 37°C for 2 h, followed by the addition of soluble CD4 (sCD4) at 200 ng/ml for 2 h. To determine IC50, BMS-378806 was added simultaneously with sCD4. After washing with buffer (20 mM Tris·HCl/500 mM NaCl/0.05% Tween 20, pH 7.5), CD4 antibody, OKT4, was added to plate for 1 h. Unbound OKT4 was removed and an anti-mouse peroxidase conjugate (Bio-Rad) was added for 1 h. After washing, 3,3′,5,5′-tetramethyl-benzidine chromogen substrate (Pierce) for peroxidase was added and the optical density was read at 450 nm.
Compound/gp120 Binding Assay. Micro BioSpin 6 columns (Bio-Rad) capable of separating small ligands from large macromolecules were used to measure the binding of [3H]BMS-378806 to gp120. Binding solution containing 25 mM Tris·HCl (pH 7.5), 125 mM NaCl, 5–200 nM gp120, and 100 nM [3H]BMS-378806 was allowed to equilibrate for 5 min, adsorbed to a Micro BioSpin 6 column, and centrifuged for 5 sec. The eluent was counted in a scintillation counter.
Selection of Drug-Resistant Variants. BMS-378806-resistant HIV-1 variants were selected (25). Selection began at a concentration twice the respective EC50, and viruses were passaged in the presence of increasing concentrations of the compound. Supernatants were collected for drug susceptibility assays, and envelope sequences were determined at select passages. Seven to eighteen independent clones were sequenced for each resistant virus.
Drug Susceptibility Assay. MT-2 cells were infected with laboratory strains at an multiplicity of infection (MOI) of 0.005, followed by incubation in the presence of serially diluted inhibitors for 4–5 days. Virus yields were quantitated by using a reverse-transcriptase assay (26). Phytohemagglutinin-activated PBMCs were infected with clinical isolates at an MOI of 0.005 and then incubated in the presence of serially diluted compounds for 6 days. The virus replication was measured by a p24 assay, and EC50 values were calculated (26).
Cell-Based Fusion Assay. An HIV-1 envelope-mediated fusion assay (27) was established by using two populations of HeLa cells. One population, the effector cells, expresses the HIV-1 gp160 protein and the tetracycline-responsive transcriptional activator (tTA, expressed from the pTET-off plasmid driven by cytomegalovirus promoter, BD Biosciences/PharMingen). The second population, the target cells, expresses the HIV-1 receptors (CD4, CCR5, and CXCR4) and a tTA-responsive luciferase reporter plasmid (pTRE-luc, BD Biosciences/PharMingen). Cell fusion allows the activation of the pTRE-luc reporter by the tTA activator and can be quantitated by monitoring luciferase activity.
To construct the pTRE-NL HIV-1 gp160 expression clone, the rev/gp160 region of the pNL4-3 proviral clone was PCR amplified and cloned into pTRE2 plasmid (BD Biosciences/PharMingen) pTRE-LAI and pTRE-mutant gp120 constructs were similarly constructed. Target cells were prepared by stably transducing HeLa cells that express CD4, CXCR4, and CCR5 receptors with the pTRE-luc plasmid. Effector cells were prepared by transiently transfecting 5 × 106 HeLa cells with 0.2 μg of pTRE-envelope construct and 1 μg of pTET-off. After an overnight incubation, the fusion process was initiated by mixing trypsinized effector and target cells in a 1:2 ratio and the mixtures were seeded in a 96-well plate at 1.5 × 104 cells per well, followed by the addition of various concentrations of compound. Luciferase activity was determined 12–24 h after mixing with the Steady-Glo Luciferase Assay system (Promega).
Generation of the Recombinant Virus Variants. The pNL4-3 plasmid was from the National Institutes of Health AIDS Reagent Repository and the pLAI plasmid was kindly provided by M. Emerman of the Fred Hutchinson Institute. Cloned viruses carrying variant envelopes were generated by site-directed mutagenesis of envelope fragments using the QuikChange site-directed mutagenesis kit (Stratagene). After confirmation by sequencing, the fragments were substituted into either pNL4-3 or pLAI and the viruses were generated by transfecting the plasmids into 293 cells. Numbering of gp120 amino acid residues is based on the sequence of the prototypic HXBc2 strain of HIV-1, according to current convention (28).
Pharmacokinetic Studies in Animals. Studies were conducted in rats, dogs, and cynomolgus monkeys after i.v. and oral administration of BMS-378806 in a solution formulation (90% polyethylene glycol 400/10% ethanol). The i.v. doses were 1 mg/kg in rats and 0.67 mg/kg in dogs and monkeys, and the oral doses were 5 mg/kg in rats and 3.4 mg/kg in dogs and monkeys. Animals (n = 3 per route) were fasted overnight, and plasma samples were collected before and after dosing for 24 h (8 h in rats). The drug concentrations in plasma were determined by liquid chromatography tandem mass spectrometry (LC/MS/MS) after protein precipitation of samples. The pharmacokinetic parameters (area under the curve, bioavailability, clearance, and half-life, etc.) were calculated from the plasma concentration time data by noncompartmental analysis using kinetica software (version 3.0, InnaPhase, Philadelphia).
Results
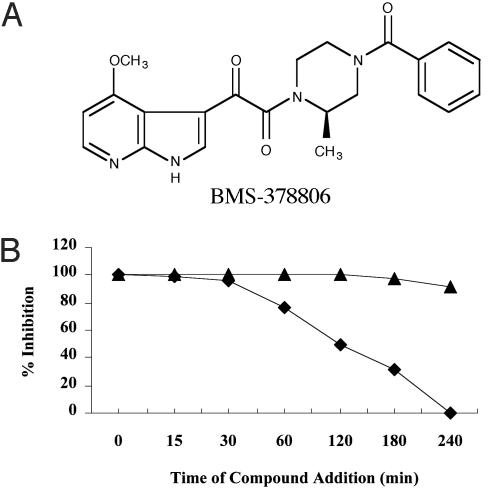
Biological Properties of BMS-378806. BMS-378806 was identified from a whole cell infection screen against HIV-1JRFL. Our initial screen hit was an indole analog with an EC50 of 0.2 μM against HIVJRFL and a CC50 of 226 μM against HeLa cells. This molecule was further optimized to improve potency, specificity and pharmacokinetic parameters. The resulting compound, BMS-378806, is a 4-methoxy-7-azaindole derivative with a molecular weight of 406.5 (Fig. 1A) and an aqueous solubility of ≈200 μg/ml (pH 8.4). BMS-378806 has excellent potency against a panel of 11 HIV-1 laboratory strains, with a median EC50 of 12 nM (range 0.9–743 nM), and is effective against viruses using the CCR5 coreceptor (R5), viruses using the CXCR4 coreceptor (X4), and R5/X4 HIV-1 strains (Table 1). The compound showed no significant cytotoxicity (CC50 > 225 μM) when assayed in 14 different cell lines (see Materials and Methods) representing tissues from multiple organs and cell types (data not shown).
Fig. 1.
(A) The structure of BMS-378806. (B) Time-of-addition assay. HeLa CD4 CCR5 cells were infected with HIV-1JRFL pseudotyped virions, encoding a luciferase reporter gene. At the times indicated, either 50 nM BMS-378806 (▪) or 1 μM nevirapine (▴) (10× EC50) was added. At 4 h, the virus was removed and only the media and compounds were put back. Antiviral activity was measured after 48 h by monitoring the luciferase activity.
Table 1. Anti-HIV-1 activity of BMS-378806 against laboratory adapted strains.
| HIV-1 | Coreceptor | EC50, nM ± SD | Host |
|---|---|---|---|
| TLAV* | X4/R5 | 0.9 ± 0.3 | MT-2/CCR5 |
| JRFL† | R5 | 1.5 ± 0.6 | PM1 |
| LAI* | X4 | 2.7 ± 1.6 | MT-2 |
| SF-162† | R5 | 3.5 ± 0.8 | PM1 |
| NL 4-3* | X4 | 4 ± 2.3 | MT-2 |
| Bal† | R5 | 8.4 ± 1.8 | Macrophage |
| Bal† | R5 | 15.5 ± 6.8 | PM1 |
| A0-18* | X4 | 23.6 ± 5.8 | MT-2 |
| SF-2* | X4 | 26.5 ± 3.5 | MT-2 |
| IIIB* | X4 | 39.4 ± 9.6 | MT-2 |
| 89.6* | X4/R5 | 75.8 ± 42.1 | MT-2/CCR5 |
| MN* | X4 | 743 ± 149 | MT-2 |
Antiviral end point, reverse transcriptase.
Antiviral end point, p24.
BMS-378806 exhibits specific anti-HIV-1 activity, because no appreciable activity is observed against HIV-2, simian immunodeficiency virus, murine leukemia virus, cytomegalovirus, respiratory syncytial virus, influenza virus, and bovine viral diarrhea virus when tested at concentrations ranging from 10 to >300 μM (data not shown). Furthermore, BMS-378806 is not inhibitory against a panel of 71 diverse cellular receptors at 10 μM (unpublished data).
Inhibition of Viral Entry. Initially, BMS-378806 was found to be inactive against HIV-1 reverse transcriptase (IC50 > 100 μM), protease (IC50 > 20 μM), and integrase (IC50 > 500 μM). To determine the antiviral target of BMS-378806, a time-of-addition experiment was performed by using a single cycle infection assay (21). HeLa cells (expressing CD4 and CCR5) were infected with HIV-1JRFL pseudotyped virions, and either 50 nM BMS-378806 or 1 μM nevirapine were added at various times after infection. Nevirapine, which inhibits reverse transcriptase at a later stage of the life cycle, retains activity even if added 4 h after infection. In contrast, a decrease in inhibition is observed when BMS-378806 is added >30 min after infection, suggesting that an early stage of viral infection is the likely target (Fig. 1B).
Because the HIV entry process involves at least three sequential stages, additional experiments were performed to further distinguish the precise target of inhibition. First, BMS-378806 was shown to inhibit the fusion of effector cells expressing the gp160 envelopes of HIV-1LAI,HIV-1NL4-3, or HIV-1JRFL with target cells expressing the CD4 receptor and CCR5/CXCR4 coreceptors with EC50 values of 2.3, 2.7, and 4.2 nM, respectively (data not shown). The fusion data, combined with results from the time-of-addition experiment, supported the hypothesis that BMS-378806 blocks replication at the stage of viral entry. A review of the data presented in Table 1 indicates that BMS-378806 inhibition is independent of the coreceptor used by the tested viral strains, suggesting that CCR5 or CXCR4 coreceptor is unlikely to be the target of BMS-378806. To further rule out that BMS-378806 interferes with coreceptor interaction, U87-CD4 cells (22), which do not express any chemokine receptors, were transfected with an expression vector encoding either CCR5 or CCR3 and then infected with an ADA-Renilla luciferase reporter virus (23). The EC50 value obtained from both sets of cells were similar, 1.9 nM (CCR5) and 3.7 nM (CCR3), confirming that the BMS-378806 inhibitory activity does not involve either the CCR5 or CCR3 coreceptor (data not shown).
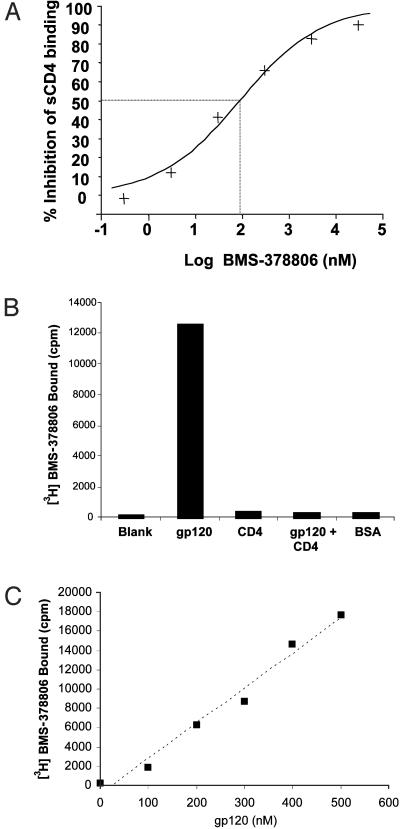
To determine whether virus binding to CD4 receptor is the compound target, an ELISA assay was established to measure the effect of BMS-378806 on gp120/CD4 interactions. Results with the HIV-1JRFL envelope demonstrated that BMS-378806 inhibits gp120/CD4 binding with an IC50 of ≈100 nM (Fig. 2A). To further elucidate how BMS-378806 interferes with the binding of gp120 to CD4 receptors, a Micro BioSpin 6 column gel filtration method was used. [3H]BMS-378806 was shown to bind selectively to gp120 as demonstrated by the coelution of [3H]BMS-378806 with gp120 from a column on centrifugation (Fig. 2B). When [3H]BMS-378806 was mixed with increasing amounts of gp120 and loaded onto individual columns, the amount of [3H]BMS-378806 eluted from the columns on centrifugation was directly proportional to the gp120 present (Fig. 2C). In contrast, when sCD4 at a 10-fold excess or a control protein, BSA at a 49-fold excess were used in place of gp120, the amount of [3H]BMS-378806 eluted was at background levels, indicating specific binding to gp120 (Fig. 2B). BMS-378806 failed to bind gp120 if the protein was prebound with sCD4 (Fig. 2B). From this series of studies, we conclude that BMS-378806 is an attachment inhibitor that interferes with gp120/CD4 interactions and that this inhibition is likely mediated through binding to the viral envelope gp120 and not the CD4 receptor.
Fig. 2.
Mode-of-action studies on BMS-378806. (A) Inhibition of gp120/CD4 binding by BMS-378806. The dose-dependent inhibition of sCD4 binding to gp120 by BMS-378806 gave an IC50 of ≈100 nM, indicating that the compound interferes with gp120/CD4 binding. (B and C) Gel filtration analysis of [3H]BMS-378806 binding to gp120. [3H]BMS-378806 was mixed with the proteins shown and then centrifuged through a Micro BioSpin 6 gel filtration column. Protein-bound radiolabel was quantitated in the eluent by liquid scintillation counting. (B) Samples contained no protein (Blank), 500 nM gp120, 3 μM CD4, 500 nM gp120 and 3 μM CD4, or 25 μM BSA. (C) Samples contained 0–500 nM gp120. The amount of [3H]BMS-378806 bound was proportional to the amount of gp120 added.
Resistant HIV-1 Variants. To study the potential of resistance development and to further define the antiviral target of BMS-378806 in infected cells, HIV-1NL4-3 and HIV-1LAI were passaged in cell culture in the presence of increasing concentrations of compound to select for resistant viruses. In general, the rate of resistance development of BMS-378806 (20 days or more) is comparable to nevirapine and lamivudine in semiquantitative parallel studies (unpublished data). The susceptibility of emerging BMS-378806-resistant viruses along with the genotype of HIV-1 variant envelopes were determined, and only the frequently encountered amino acid substitutions, along with the envelope domain in which they reside were listed (9) (Table 2). These substitutions are I595F, M475I, M434I/V, K655E, R350K, S440R, V68A, D185N, and M426L. Interestingly, I595F and K655E reside in gp41, whereas all other substitutions span the entire gp120 region. In particular, M426L and M475I are located at or near the gp120/CD4 contact sites (13, 14). The M434I and M475I substitutions appear to play a key role in BMS-378806 resistance development, because they were selected in two independent experiments using HIV-1LAI and HVI-1NL4-3, respectively. In a sequential selection of BMS-378806 resistant variants (HIV-1LAI, experiment 1), resistance levels increased >60-fold as drug concentration was raised from 96 to 384 nM (Table 2). This decreased susceptibility appeared to be correlated with the emergence of M434I (from 0% to 62.5%). Regarding M426L, it appeared rarely in BMS-378806-selected variants, but occurred frequently when related analog compounds were used in the selection process (data not shown).
Table 2. Phenotypes and genotypes of BMS-378806-resistant viruses.
| HIV-1 | Experiment | Selection, nM | Fold resistance* | Substitutions | Frequency | Domain |
|---|---|---|---|---|---|---|
| NL 4-3 | 1 | 96 | 20 | M475I | 8/10 | C5 |
| 2 | 100 | 218 | M475I | 5/18 | C5 | |
| R350K/M475I | 4/18 | C3/C5 | ||||
| D185N/R350K/M475I | 2/18 | V2/C3/C5 | ||||
| LAI | 1 | 96 | 31 | I595F | 6/7 | gp41 |
| 384 | 1,941 | M434I | 1/8 | C4 | ||
| I595F | 2/8 | gp41 | ||||
| M434I/I595F | 4/8 | C4/gp41 | ||||
| I595F/K655E | 1/8 | gp41/gp41 | ||||
| 2 | 33 | 157 | M434I/V | 8/16 | C4 | |
| S440R | 2/16 | C4 | ||||
| V68A/M434I | 1/16 | C1/C4 | ||||
| V68A/S440R | 1/16 | C1/C4 | ||||
| M426L/K655E | 1/16 | C4/gp41 | ||||
| M434I/K655E | 1/16 | C4/gp41 | ||||
| S440R/K655E | 2/16 | C4/gp41 |
Major substitutions are shown in bold.
Fold changes in susceptibility, significant at the ≤0.05 level by an unpaired t test.
To further elucidate the impact of these BMS-378806-selected envelope substitutions on viral resistance, the changes were incorporated into HIV-1 envelopes and the resulting recombinant viruses were used to evaluate their susceptibility to BMS-378806. Individually, M426L, M434I, and M475I substitutions reduced the susceptibility of HIV-1LAI or HIV-1NL4-3 by 116-, 4-, and 40-fold, respectively (Table 3). In addition, double mutant viruses were generated for HIV-1LAI harboring M434I and one of the two secondary substitutions (V68A or I595F). Although V68A or I595F individually had some effect on viral susceptibility, the presence of either one of these two substitutions enhance the resistance levels of M434I by an additional 19- and 125-fold, respectively (Table 3). Thus, the enhanced resistance level of the M434I/I595F virus contributes to the highly resistance phenotype of HIVLAI selected by BMS-378806 at a concentration of 384 nM (Table 2). Data to date show that the V68A, M426L, M434I, M475I, and I595F changes in viral envelope all confer BMS-378806 resistance to the recombinant HIV-1 variants to various degrees, confirming that the viral envelope is the target of inhibitor binding.
Table 3. Phenotypes of recombinant viruses carrying in vitro mutagenized envelopes.
| Envelope | Substitutions | EC50, nM ± SD | Fold resistance* |
|---|---|---|---|
| LAI | Wild type | 4 ± 1 | 1 |
| V68A | <15 | <4 | |
| V68A/M434I | 309 ± 72 | 77 | |
| M426L | 462 ± 158 | 116 | |
| M434I | 17 ± 2 | 4 | |
| M434I/I595F | 1,990 ± 70 | 498 | |
| I595F | 32 ± 10 | 8 | |
| NL4-3 | Wild type | 67 ± 4 | 1 |
| M475I | 2,689 ± 304 | 40 | |
| R350K | 38 ± 4 | 0.6 | |
| R350K/M475I | 1,028 ± 296 | 15 |
Major substitutions are shown in bold.
Fold resistance, EC50 for resistant variants/EC50 wild-type strains.
Anti-HIV-1 Spectrum. Because of the inherent heterogeneity of the HIV-1 envelope, it is critical to evaluate the antiviral activity of BMS-378806 against a large panel of HIV-1 clinical isolates to determine whether a drug targeting viral attachment would provide a broad coverage. This is particularly important in light of the results in Table 1, in which HIV-1MN laboratory strain was not efficiently inhibited by BMS-378806 (EC50 = 743 nM).
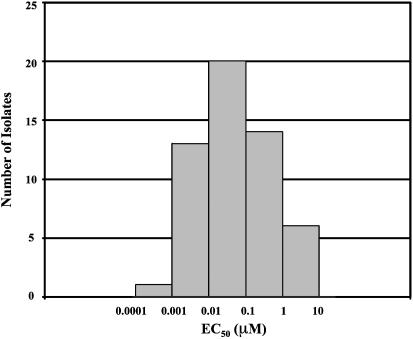
HIV-1 isolates can be classified into distinct subtypes. Evaluation of 53 strains of the B subtype showed that BMS-378806 exhibited a wide spectrum of activity with a median EC50 of 0.04 μM (Fig. 3). Of the 42 clinical isolates and 11 laboratory strains (see Table 1) examined, the majority (62%) showed EC50 of <0.1 μM (Fig. 3). Six virus strains showed poor susceptibility or were refractory to BMS-378806 at the highest concentrations tested (1–10 μM). The remaining viruses tested showed intermediate susceptibility (0.1 to 1 μM). A panel of 41 non-B subtype clinical isolates was also evaluated. As summarized in Table 4, BMS-378806 exhibited decreased, but still significant activity against the subtype C viruses, low activity against viruses from subtypes A and D, and poor or no activity vs. subtypes E, F, G, and Group O viruses (Table 6, with data for each isolate, is published as supporting information on the PNAS web site, www.pnas.org). However, the number of viruses tested in these latter groups remain limited.
Fig. 3.
Susceptibility histogram for subtype B clinical isolates and laboratory strains. BMS-378806 susceptibility was plotted against 42 clinical isolates and 11 laboratory strains of the HIV-1 B subtype. The x axis indicates EC50 (in μM), with the left boundary of each bar listing the lower limit and the right boundary showing the upper limit of EC50. The y axis represents the number of isolates that falls within a particular range of EC50. The median EC50 is 0.04 μM.
Table 4. Anti-HIV potency of BMS-378806 against 83 clinical isolates from different envelope subtypes.
| Group | Subtype | No. of isolates | Median EC50 (range, nM) |
|---|---|---|---|
| M | A | 5 | 1,132 (51.3 to >5,000) |
| B | 42 | 61.5 (1 to >10,000) | |
| C | 12 | 243 (11.8-749) | |
| D | 10 | 2,183 (45.8 to >10,000) | |
| E | 6 | (>2,000 to >10,000) | |
| F | 3 | (214 to >5,000) | |
| G | 3 | (>2,000-4,290) | |
| O | 2 | (>2,000) | |
| Total | 83 |
Pharmacokinetic and Safety Profiles. Whether a wide spectrum of susceptibilities will be problematic for attachment inhibitors remains to be seen. Efficacy will be determined to a great extent by the drug exposure levels achievable in man. Because the efficacy of antiretroviral agents can be impacted significantly by serum protein binding, the effect of human serum on the anti-HIV activity of BMS-378806 was assessed by infecting either HIV-1LAI or HIV-1NL4-3 strain in MT-2 cells in the presence of 40% human serum. Results showed that the presence of human serum only marginally increased the EC50 of BMS-378806 by 1.2-fold (data not shown). In agreement, the extent of protein binding in human plasma was determined to be relatively low at 73%. Together, these results suggested that human serum will likely exert a minimal effect on the anti-HIV-1 activity of BMS-378806.
Oral bioavailability of BMS-378806 was assessed in rats, dogs, and cynomolgus monkeys, and determined to be 19%, 77%, and 24%, respectively (Table 5). Species differences in oral absorption likely contributed to different bioavailabilities across species. The steady-state volume of distribution ranged from 0.4 to 0.6 liters/kg, suggesting that the compound is distributed outside plasma. BMS-378806 did not cross the blood–brain barrier to any appreciable extent. The total body clearance relative to the hepatic blood flow was intermediate in rats and low in dogs and monkeys (Table 5). The terminal half-life after i.v. administration was 0.3, 1.2, and 0.9 h in rats, dogs, and monkeys, respectively. However, the apparent terminal half-life after oral administration was significantly longer in the rat and monkey (2.1 and 6.5 h, respectively) than that after i.v. dosing, suggesting prolonged oral absorption. In addition, in vitro metabolism studies demonstrated that the compound was metabolized by multiple cytochrome P450 enzymes and had low potential for inhibiting these major human drug-metabolizing enzymes.
Table 5. Pharmacokinetic parameters of BMS-378806 in rats, dogs, and monkeys.
| Parameter | Rat | Dog | Monkey |
|---|---|---|---|
| IV/oral dose, mg/kg | 1/5 | 0.67/3.4 | 0.67/3.4 |
| Bioavailability, % | 19 | 77 | 24 |
| Oral Cmax, μM | 0.22 | 7.4 | 0.51 |
| Oral AUC0-∞, μM × h | 1.3 | 19 | 3.2 |
| Total body clearance, ml/min per kg | 32 | 5.2 | 10 |
| Steady-state volume of distribution, liters/kg | 0.56 | 0.47 | 0.39 |
| IV terminal half-life, h | 0.3 | 1.2 | 0.9 |
| Apparent oral terminal half-life, h | 2.1 | 1.4 | 6.5 |
In a 2-week exploratory drug safety study in rats, BMS-378806 was well tolerated when given orally to rats for 2 weeks at daily doses of up to 100 mg/kg. In addition, BMS-378806 was not mutagenic in an Ames reverse-substitution assay (data not shown).
Discussion
Early efforts to block HIV-1 entry to cells focused on using sCD4 to inhibit virus infection (29, 30). Although sCD4 demonstrated efficacy against many laboratory strains, it exhibited poor activity against primary isolates (31), which may have contributed to disappointing results in clinical trials (31, 32). PRO-542 (a CD4-IgG fusion protein) was shown to be effective in neutralizing many clinical HIV-1 strains in culture (15, 33) and is efficacious in the clinics (16, 34). However, this recombinant fusion protein requires i.v. administration, which may limit its clinical application. Recently, a 27-aa CD4 mimic, CD4M33, was shown to bind to gp120 and inhibit HIV-1 infection in vitro (35). The clinical development potential of this peptide awaits future studies. BMS-378806 is a small molecule inhibitor that blocks the HIV entry process. The compound specifically inhibits gp120 binding to cellular CD4 receptors, functioning independently of viral coreceptors. The pharmacokinetic and pharmaceutic characteristics of BMS-378806 support an oral formulation in man, and initial toxicology studies raised no safety concerns. As such, this class of attachment inhibitors appears suitable for advancement into clinical development.
HIV-1 envelope is a heterogeneous protein with variable sequences exposed on the outer surface of virions. Therefore, the finding of a specific and effective small molecule inhibitor of gp120 was somewhat unexpected. However, cumulative results from biochemical and genetic studies described here strongly suggest that BMS-378806 indeed targets gp120 and interferes with CD4 binding. Further studies using a fluorescence quenching method showed that BMS-378806 binds directly to gp120 at a 1:1 stoichiometry (unpublished data). These studies establish the feasibility of targeting the HIV-1 gp120 envelope with a small molecule compound and support the envelope protein as a viable target for a new drug class. Of particular importance is that drugs targeting the envelope are effective against strains resistant to existing classes of drugs.
Because of the diversity of envelope glycoproteins, it is not surprising that considerable variability was observed in the susceptibility of virus subtypes to BMS-378806. The most susceptible HIV-1 strains observed belong to subtype B (with a median EC50 of 0.04 μM) (Fig. 3). This finding is probably due to our utilization of a subtype B strain (HIV-1JRFL) in the initial screening process. BMS-378806 does inhibit other subtypes, but at a lower potency (Table 4). Enfuvirtide and coreceptor inhibitors also exhibit a range of susceptibilities when tested against various viruses (36, 37). New structural information on the precise gp120 binding site of BMS-378806 and a knowledge of the structure-activity relationship of this series of compounds will assist in the understanding of the basis of anti-HIV spectrum. The information should also allow the design of additional attachment inhibitors with enhanced potency and an expanded spectrum.
The HIV entry process depends on multiple conformational changes of the envelope protein and, together with the rapid immune evasion by HIV-1, suggests that the viral envelope is a flexible and adaptable protein. Consequently, resistance development may be an issue for compounds that target the envelope. The initial in vitro drug selection studies showed that BMS-378806 developed resistance in ≈20 days under our experimental conditions. This rate of resistance development was similar to that of the reverse transcriptase inhibitors, nevirapine and lamivudine, observed from parallel experiments (data not shown). Because HIV-1 envelope is a heterogeneous protein, it may in part explain the relatively rapid rate of resistance selection by BMS-378806. Genetic analysis of the drug selected viruses revealed that amino acid substitutions acquired during selection were mapped to regions spanning the entire HIV envelope. No changes were identified in the overall HIV-1 sequences outside the envelope region of the drug selected viruses after sequencing one entire BMS-378806-resistant HIV-1 strain (data not shown). Most significantly, M426L and M475I substitutions, located at or near gp120/CD4 contact sites, conferred high levels of resistance to the in vitro mutagenized HIV-1 variants, suggesting that the CD4 binding pocket of gp120 is the antiviral target. The M434I and other secondary changes (V68A and I595F) also affected drug susceptibility of recombinant viruses, presumably by influencing the gp120 conformation. Moreover, two of the selected substitutions, I595F and K655E, were situated at the gp41 region. It is interesting to note that the same I595 residue has been implicated in the development of sCD4 sensitivity (38). The reasons why these gp41 substitutions were selected by BMS-378806 and the impact these changes have on envelope conformation await further studies. We also noted that the selected gp120 substitutions varied depending on the viral background. This phenomenon was observed for HIV-1 protease inhibitors as well (39). Namely, M475I was the major change occurring in HIV-1NL4-3, and M434I was the major change in HIV-1LAI. It is likely that the collective interactions of these selected substitutions with other envelope residues determine the overall drug susceptibility. However, it is important to note that the available crystal structures are post-CD4 binding, and we do not know the actual conformation of gp120 targeted by BMS-378806. Finally, results from resistance studies, in conjunction with the [3H]BMS-378806/gp120 binding data, support the envelope as the target site of the compound. The mapping of the BMS-378806 resistance to the viral envelope also demonstrated that BMS-378806 functions as an inhibitor of the envelope in HIV-1-infected cells.
BMS-378806 is effective against variants resistant to HIV-1 protease and reverse transcriptase inhibitors (unpublished data). In reciprocal experiments, BMS-378806 resistant HIV strains remain susceptible to these two classes of drugs as well. Therefore, BMS-378806 may represent an important therapeutic option, particularly for patients who harbor viruses resistant to existing antiretroviral agents. In summary, results described here indicate that small molecule inhibitors of HIV-1 envelope could potentially be developed into a new class of antiretroviral drugs. The HIV-1 gp120 envelope represents a recently discovered phase of the viral life cycle that can be effectively targeted with small molecule compounds. Further studies with this class of inhibitors may also provide us with a greater understanding of the HIV-1 envelope structure and the HIV entry process.
Supplementary Material
Acknowledgments
We thank Dr. R. Levitz for securing some clinical isolates, Dr. D. Dischino for preparing [3H]BMS-378806, P. Poundstone for sequencing some isolates, and C. Hendricks for help preparing the manuscript.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviation: sCD4, soluble CD4 protein; R5, virus using the CCR5 coreceptor; X4, virus using the CXCR4 coreceptor; CC50, concentration for 50% reduction of cell growth.
See commentary on page 10581.
Footnotes
Reynes, J., Rouzier, R., Kanouni, T., Baillat, V., Baroudy, B., Keung, A., Hogan, C., Markowitz, M. & Laughlin, M. (2002) 9th Conference on Retroviruses and Opportunistic Infections, Seattle, WA, abstr. 1.
References
- 1.Cohen, O. J. & Fauci, A. S. (2001) Adv. Intern. Med. 46, 207–246. [PubMed] [Google Scholar]
- 2.Volberding, P. (1999) AIDS 13, S1–S9. [PubMed] [Google Scholar]
- 3.Wegner, S. A., Brodine, S. K., Mascola, J. R., Tasker, S. A., Shaffer, R. A., Starkey, M. J., Barile, A., Martin, G. J., Aronson, N., Emmons, W. W., et al. (2000) AIDS 14, 1009–1015. [DOI] [PubMed] [Google Scholar]
- 4.Little, S. J., Holte, S., Routy, J. P., Daar, E., Markowitz, M., Collier, A. C., Koup, R. A., Mellors, J. W., Connick, E., Conway, B., et al. (2002) N. Engl. J. Med. 347, 385–394. [DOI] [PubMed] [Google Scholar]
- 5.LaBranche, C. C., Galasso, G., Moore, J. P., Bolognesi, D., Hirsch, M. S. & Hammer, S. M. (2001) Antiviral Res. 50, 95–115. [DOI] [PubMed] [Google Scholar]
- 6.Pierson, T. C. & Doms, R. W. (2003) Immunol. Lett. 85, 113–118. [DOI] [PubMed] [Google Scholar]
- 7.Chan, D. C. & Kim, P. S. (1998) Cell 93, 681–684. [DOI] [PubMed] [Google Scholar]
- 8.Wyatt, R. & Sodroski, J. (1998) Science 280, 1884–1888. [DOI] [PubMed] [Google Scholar]
- 9.Starcich, B. R., Hahn, B. H., Shaw, G. M., McNeely, P. D., Modrow, S., Wolf, H., Parks, E. S., Parks, W. P., Josephs, S. F., Gallo, R. C., et al. (1986) Cell 45, 637–648. [DOI] [PubMed] [Google Scholar]
- 10.Robertson, D. L., Anderson, J. P., Bradac, J. A., Carr, J. K., Foley, B., Funkhouser, R. K., Gao, F., Hahn, B. H., Kalish, M. L., Kuiken, C., et al. (1999) HIV-1 Nomenclature Proposal (Theoretical Biology and Biophysics Group, Los Alamos National Laboratory, Los Alamos, NM).
- 11.Robertson, D. L., Anderson, J. P., Bradac, J. A., Carr, J. K., Foley, B., Funkhouser, R. K., Gao, F., Hahn, B. H., Kalish, M. L., Kuiken, C., et al. (2000) Science 288, 55–56. [DOI] [PubMed] [Google Scholar]
- 12.Hu, D., Dondero, T. J., Mastro, T. D. & Gayle, H. D. (1998) in AIDS and Other Manifestations of HIV Infection: 1998, ed. Wormser, G. P. (Lippincott-Raven, Philadelphia), pp. 27–40.
- 13.Kwong, P. D., Wyatt, R., Mcajeed, S., Robinson, J., Sweet, R. W., Sodroski, J. & Hendrickson, W. A. (2000) Structure (London) 8, 1329–1339. [DOI] [PubMed] [Google Scholar]
- 14.Kwong, P. D., Wyatt, R., Robinson, J., Sweet, R. W., Sodroski, J. & Hendrickson, W. A. (1998) Nature 393, 648–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Trkola, A., Pomales, A. B., Yuan, H., Korber, B., Maddon, P. J., Allaway, G. P., Katinger, H., Barbas, C. F., III, Burton, D. R., Ho, D. D., et al. (1995) J. Virol. 69, 6609–6617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shearer, W. T., Israel, R. J., Starr, S., Fletcher, C. V., Wara, D., Rathore, M., Church, J., DeVille, J., Fenton, T., Graham, B., et al. (2000) J. Infect. Dis. 182, 1774–1779. [DOI] [PubMed] [Google Scholar]
- 17.Lalezeri, J. P., Henry, K., O'Hearn, M., Montaner, J. S. G., Piliero, P., Trottier, B., Walmsley, S., Cohen, C., Kuritzkes, D. R., Eron, J. J., Jr., et al. (2003) N. Engl. J. Med. 348, 2175–2185. [DOI] [PubMed] [Google Scholar]
- 18.Kilby, J. M., Lalezari, J. P., Eron, J. J., Carlson, M., Cohen, C., Arduino, R. C., Goodgame, J. C., Gallant, J. E., Volberding, P., Murphy, R. L., et al. (2002) AIDS Res. Hum. Retroviruses 18, 685–693. [DOI] [PubMed] [Google Scholar]
- 19.Kilby, J., Hopkins, S., Venetta, T. M., DiMassimo, B., Cloud, G. A., Lee, J. Y., Alldredge, L., Hunter, E., Lambert, D., Bolognesi, D., et al. (1998) Nat. Med. 4, 1302–1307. [DOI] [PubMed] [Google Scholar]
- 20.Weislow, O. S., Kiser, R., Fine, D. L., Bader, J., Shoemaker, R. H. & Boyd, M. R. (1989) J. Natl. Cancer Inst. 81, 577–586. [DOI] [PubMed] [Google Scholar]
- 21.Donzella, G. A., Schols, D., Lin, S. W., Este, J. A., Nagashima, K. A., Maddon, P. J., Allaway, G. P., Sakmar, T. P., Henson, G., De Clercq, E. & Moore, J. P. (1998) Nat. Med. 4, 72–77. [DOI] [PubMed] [Google Scholar]
- 22.Hill, C. M., Deng, H., Unutmaz, D., Kewalramani, V. N., Bastiani, L., Gorny, M. K., Zolla-Pazner, S. & Littman, D. R. (1997) J. Virol. 71, 6296–6304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blair, W. & Spicer, T. P. (2003) U.S. Patent 0013078 A1.
- 24.Moore, J. P. (1989) AIDS 3, 155–163. [DOI] [PubMed] [Google Scholar]
- 25.Gong, Y. F., Robinson, B. S., Rose, R. E., Deminie, C., Spicer, T. P., Stock, D., Colonno, R. J. & Lin, P. F. (2000) Antimicrob. Agents Chemother. 44, 2319–2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johnson, V. A. & Byington, R. E. (1990) Infectivity Assay (Virus Yield Assay) (Stockton, New York).
- 27.Nussbaum, C., Broder, C. & Berger, E. A. (1994) J. Virol. 68, 5411–5422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Korber, B., Foley, F., Kuiken, C., Pillai, S. & Sodroski, J. (1998) in Human Retroviruses and AIDS, eds. Korber, B., Kuiken, C., Foley, B., Hahn, B. H., McCutchan, F. E., Mellors, J. W. & Sodroski, J. (Los Alamos National Laboratory, Los Alamos, NM), pp. III-102–III-103.
- 29.Smith, D. H., Byrn, R. A., Marsters, S. A., Gregory, T., Groopman, J. E. & Capon, D. J. (1987) Science 238, 1704–1707. [DOI] [PubMed] [Google Scholar]
- 30.Hussey, R. E., Richardson, N. E., Kowalski, M., Brown, N. R., Chang, H. C., Siliciano, R. F., Dorfman, T., Walker, B., Sodroski, J. & Reinherz, E. L. (1988) Nature 331, 78–81. [DOI] [PubMed] [Google Scholar]
- 31.Daar, E. S., Li, X. L., Moudgil, T. & Ho, D. D. (1990) Proc. Natl. Acad. Sci. USA 87, 6574–6578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schacker, T., Coombs, R. W., Collier, A. C., Zeh, J. E., Fox, I., Alam, J., Nelson, K., Eggert, E. & Corey, L. (1994) J. Infect. Dis. 169, 37–40. [DOI] [PubMed] [Google Scholar]
- 33.Allaway, G. P., Davis-Bruno, K. L., Beaudry, G. A., Garcia, E. B., Wong, E. L., Ryder, A. M., Hasel, K. W., Gauduin, M. C., Koup, R. A., McDougal, J. S., et al. (1995) AIDS Res. Hum. Retroviruses 11, 533–539. [DOI] [PubMed] [Google Scholar]
- 34.Jacobson, J. M., Lowy, I., Fletcher, C. V., O'Neill, T. J., Tran, D. N., Ketas, T. J., Trkola, A., Klotman, M. E., Maddon, P. J., Olson, W. C. & Israel, R. J. (2000) J. Infect. Dis. 182, 326–329. [DOI] [PubMed] [Google Scholar]
- 35.Martin, L., Stricher, F., Misse, D., Sironi, F., Pugniere, M., Barthe, P., Prado-Gotor, R., Freulon, I., Magne, X., Roumestand, C., et al. (2003) Nat. Biotechnol. 21, 71–76. [DOI] [PubMed] [Google Scholar]
- 36.McCombie, S. W., Tagat, J. R., Vice, S. F., Lin, S.-I., Steensma, R., Palani, A., Neustadt, B. R., Baroudy, B. M., Strizki, J. M., Endres, M., et al. (2003) Bioorg. Med. Chem. Lett. 13, 567–571. [DOI] [PubMed] [Google Scholar]
- 37.Labrosse, B., Labernardiere, J.-L., Dam, E., Trouplin, V., Skrabal, K., Clavel, F. & Mammano, F. (2003) J. Virol. 77, 1610–1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moore, J. P., Burkly, L. C., Connor, R. I., Cao, Y., Tizard, R., Ho, D. D. & Fisher, R. A. (1993) AIDS Res. Hum. Retroviruses 9, 529–539. [DOI] [PubMed] [Google Scholar]
- 39.Rose, R. E., Gong, Y. F., Greytok, J. A., Bechtold, C. M., Terry, B. J., Robinson, B. S., Alam, M., Colonno, R. J. & Lin, P. F. (1996) Proc. Natl. Acad. Sci. USA 93, 1648–1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.