Abstract
Progressive rod–cone degeneration (prcd) is the most widespread hereditary retinal disease leading to blindness in dogs and phenotypically is the canine counterpart of retinitis pigmentosa (RP) in humans. In previous efforts to identify the genetic locus for prcd, the canine homologs for many of the genes causally associated with RP in humans, such as RHO, PDE6B, and RDS/peripherin, have been excluded. In parallel with a recent undertaking to establish a framework map of the canine genome, multiple prcd-informative pedigrees have been typed with a panel of more than 100 anchor loci and microsatellite-based markers. Identification of a linkage group flanking prcd ([TK1, GALK1, prcd]–[MYL4, C09.173, C09.2263]–RARA–C09.250–C09.474–NF1) localizes prcd close to the centromeric end of canine chromosome 9 (CFA9), and excludes RARA as a candidate gene. The conserved synteny of this region of CFA9 and distal human chromosome 17q establishes the potential locus homology of prcd in the dog with RP17, a human retinitis pigmentosa locus for which no gene has yet been identified. Assignment of the prcd disease locus to an identified canine autosome represents a powerful application of the developing canine linkage map in medical genetics. The usefulness of this approach is further demonstrated by identification of the correspondence of the prcd interval to homologous human and mouse chromosomal regions. The rapid progress that is now occurring in the field of canine genetics will expedite the identification of the genes underlying many of the inherited traits and diseases that make the dog a unique asset for the study of mammalian traits.
The domesticated dog, Canis familiaris, exhibits a striking range of phenotypes that are clearly hereditary. These range from relatively simple traits, such as variation in coat color (1) and numerous single gene disorders (2–4), through a range of well described clinical disorders and other phenotypes exhibiting complex inheritance (3–10). In addition, there is the diverse repertoire of morphological and/or behavioral characteristics that define and distinguish the numerous specific breeds of dog. In many cases these phenotypes are both of interest in and of themselves and have such compelling similarity to recognizable human traits that they promise unique insights into the responsible genetic and metabolic mechanisms. However, despite the enormous range of genetic traits to investigate, progress in mapping the corresponding loci has been hampered, until recently, by the paucity of information about the canine genome. This problem has been compounded by the difficulties in karyotyping the dog genome, which has a large number (38 pairs of autosomes, X, Y) of small, similar, and mostly acrocentric chromosomes (11–14). With the exception of a few X-linked disease phenotypes (see refs. 15 and 16 for examples), so far no heritable trait has been mapped to an identified canine chromosome.
Although the canine genome map is still very sparse (17–19) compared with that of well mapped species, recent efforts to develop a framework linkage map of the canine genome (17, 19), including both gene-specific (type I, or anchor locus) and anonymous (type II) microsatellite markers, enable genomic screening by linkage analysis to be undertaken in informative canine pedigrees. Simultaneously, recent improvements in high resolution canine cytogenetics and chromosome mapping by fluorescent in situ hybridization (FISH) have further advanced mapping efforts in the dog by permitting assignment of anchor loci and linkage groups to defined chromosomes (12, 13, 20). Thus, it is now feasible to undertake a comprehensive search by linkage analysis for the loci controlling heritable canine traits, and assign such loci to defined canine linkage groups and chromosomal regions. Furthermore, it is possible, by recognition of interspecies conservations of synteny, to determine the corresponding regions of the human and mouse genomes to which such traits map.
Progressive rod–cone degeneration (prcd) is a canine retinal degeneration inherited as an autosomal recessive trait (21). It is one of several diseases recognized clinically and collectively as progressive retinal atrophy (PRA), the canine phenotypic equivalent of retinitis pigmentosa (RP) in humans. Because of its clinical similarity to RP, prcd has been widely studied as a model of the human disease (21–24). The prcd phenotype is that of a degenerative disorder, in which rods and then cones degenerate both structurally and functionally after apparently normal postnatal development. For this reason it is classified as a late onset disorder, indicating that the clinical disease is not apparent until early adolescence or early adulthood (25).
Classical genetic studies established that several allelic forms of prcd occur in different canine populations (25). Candidate gene studies have excluded several of the known RP loci, such as the β-subunit gene for retinal cGMP phosphodiesterase (PDE6B), opsin, transducin α1, and RDS/peripherin (26–28). Until now, however, there has been no information regarding the map location of prcd. As a result, little is known about potential candidates or to which, if any, human RP locus prcd might correspond.
As part of an investigation into prcd, three-generation prcd-informative pedigrees were developed. These pedigrees were constructed purposefully to be as genetically polymorphic as possible, yet remain fully informative for prcd, and to achieve the largest informative sibships. This construction made them simultaneously attractive for canine genome mapping studies, and thus they were used concurrently for both purposes. Genetic mapping studies have identified a linkage group flanking both sides of the prcd locus, with placement of prcd close to the centromeric end of canine chromosome 9 (CFA9). Because this region of CFA9 corresponds to distal human chromosome 17q (29), conservation of synteny strongly suggests that prcd in the dog is the locus homolog of RP17, a human RP locus for which no gene has yet been identified (30, 31).
METHODS
Animals.
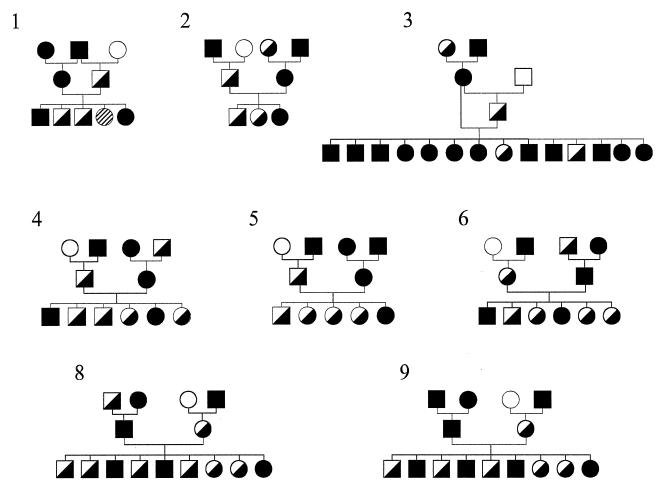
The prcd strain of dogs is maintained as part of an National Eye Institute-sponsored project (EY06855, “Models of Hereditary Retinal Degeneration”) at the Retinal Disease Studies Facility (RDSF) in Kennett Square, Pennsylvania. This strain of dogs derives from the original research colony of purebred miniature poodles in which the phenotype and inheritance of prcd were originally characterized (25, 32). To generate informative pedigrees for this study, prcd-affected dogs were bred to homozygous normal unrelated miniature poodles, beagles, and beagle-crossbred dogs, and their heterozygous offspring were then backcrossed to prcd-affected dogs to yield litters segregating for the prcd phenotype. As depicted in Figs. 1 and 2, the nine related three-generation canine families analyzed yielded 70 prcd-informative progeny; this represents a subset of the extended pedigree of the breeding colony. DNA isolated from blood and tissue samples from dogs in these families were tested in the present study.
Figure 1.
Pedigrees of prcd-informative three-generation families 1–6, 8, and 9, used in linkage studies. Circles represent females, squares represent males, open symbols indicate homozygous normal at prcd locus, solid symbols indicate prcd-affected, and half-filled symbols indicate prcd heterozygous. The hatched symbol in family 1 indicates a sibling for whom prcd phenotype could not be reliably ascertained. Although shown as separate pedigrees, these families together form part of a much larger prcd colony.
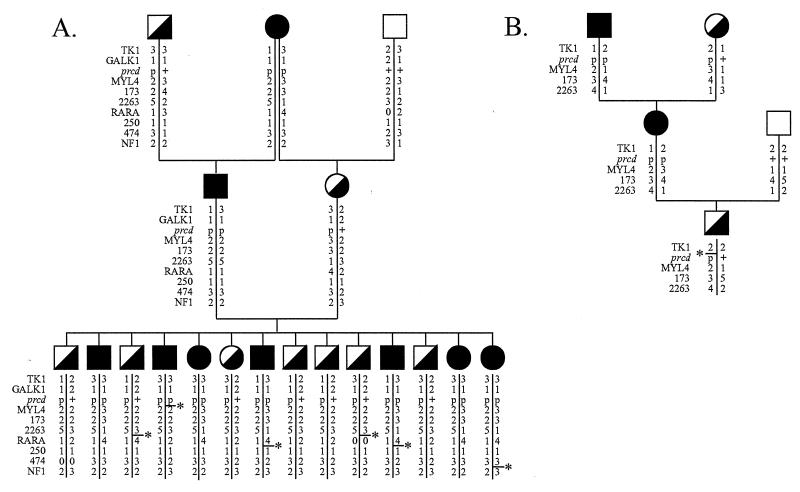
Figure 2.
(A) Pedigree of prcd-informative family 7, with haplotype data. The order of loci is according to results of two-point linkage analyses in the present study. For the prcd locus, p indicates disease allele and + indicates wild-type allele. For MYL4, allele numbers are assigned as described in the text. For all other loci, allele numbers were assigned according to ref. 19. Loci 173, 2263, 474, and 250 represent microsatellite markers C09.173, C09.2263, C09.474, and C09.250, respectively. Haplotypes demonstrating recombination events are indicated (—∗), although indicated position does not always correspond to the only possible site of the recombination. (B) Pedigree of sire of F2 litter in family 3, with haplotypes indicating that this individual received a TK1–prcd–MYL4 recombinant chromosome from his dam. Although this recombination can equally be interpreted to have taken place between TK1 and prcd, or between prcd and MYL4, the haplotypes also indicate that two different TK1 alleles are segregating in phase with prcd in this dog’s pedigree.
Diagnosis of Phenotype.
Ascertainment of prcd phenotype relied on a combination of ophthalmoscopic, electrophysiological, and retinal morphological examinations using previously published diagnostic criteria for the disease (25, 32). In dogs maintained to adulthood, initial diagnostic assignment was based on electroretinography at a minimum age of 1 year, when characteristic electroretinographic abnormalities are present in prcd-affected dogs from this colony (25). This initial diagnostic assignment was relied upon only for selection of potential breeding animals; in all dogs typed for informative pedigrees, prcd phenotype was confirmed by indirect ophthalmoscopy (for dogs over 4 years of age), retinal morphologic examination, or both. For all dogs not maintained to adulthood, diagnosis was based on retinal morphologic examination at a minimum age of 14 weeks.
Electroretinograms (ERGs) were recorded from halothane-anesthetized dogs, to stimuli and under conditions designed to enable separate evaluation of rod-mediated and cone-mediated response components. The ERG stimuli were controlled for intensity, color, and rate of presentation of 10-μsec light flashes from a Grass strobe white light source, delivered to the cornea by means of a fiber optic light guide (25, 33). For morphologic assessment of disease, eyes were fixed in mixed aldehyde solutions and embedded in an epoxy resin (33). One-micrometer sections of prcd-affected retinas demonstrate characteristic and diagnostically reliable morphologic changes by 14 weeks of age (34).
DNA Extraction.
Genomic DNA was extracted either from aliquots of fresh, or frozen and thawed, citrated whole blood samples or from frozen samples of splenic tissue, as described previously (26, 35).
Microsatellite Typing.
Microsatellites were amplified as described previously, omitting the Hot Start protocol (29, 36–38). PCR products were separated by electrophoresis on denaturing (4–6%) polyacrylamide gels (PAGE) at 55°C and were visualized by autoradiography or by fluorescence on an Applied Biosystems 377 fluorescence DNA sequencer. Genotypes determined by autoradiography were scored independently by two individuals and entered into a consensus database; disputed genotypes were omitted. All microsatellite primers have been reported previously (19, 29, 36–39).
MYL4 Polymorphism.
The human atrial myosin alkali light chain 1 gene (MYL4, GenBank accession nos. M24121 and J03954) maps to human chromosomal region 17q21–qter (ref. 40; see also the Genome Data Base at The John Hopkins University, http://www.gdb.org). Because of our initial evidence of homology between this human chromosomal segment and the prcd-linked canine chromosomal region (see below), we cloned and sequenced a 1.5-kb fragment of the canine homologue of MYL4. Genomic DNA was amplified by using primers MYL4–2, 5′-CATTGTTTGACCGGACCCCGACTGG-3′, and MYL4–4, 5′-CCTTGTTGCGGGAAATGTGCTGC-3′ and 2.5 mM MgCl2 for 40 cycles of 94°C for 30 sec, 65°C for 1 min, and 72°C for 2 min. Two restriction fragment length polymorphisms (RFLPs) have been identified in this gene fragment. Digestion with BstNI yields six nonpolymorphic and one polymorphic (140-bp) fragments. When the polymorphic site is present (allele 2) the 140-bp fragment is digested to 120- and 20-bp fragments. Absence of the polymorphic site constitutes allele 1. Digestion of the 1.5 kb-amplified fragment with BsrI generates two nonpolymorphic and one polymorphic (340-bp) fragments. Presence of the BsrI polymorphic site (allele 4) allows cleavage of the 340-bp fragment into 250- and 90-bp fragments. Absence of the polymorphic site constitutes allele 3. Digestion products were analyzed on 6% PAGE.
Linkage Analysis.
Using the linkage package of programs in refs. 41 and 42, we undertook two-point linkage analysis (43) between prcd and each marker and between each pair of markers. The disease trait was coded as an autosomal recessive trait with full penetrance and no phenocopy (41–43). Because we had prior knowledge of the actual prcd genotypes for all parental and grandparental dogs (based on previous breeding studies), prcd was also coded as an allele numbers locus. Three-point analyses were also run on selected subsets of loci.
Subsequently, the data were displayed on a pedigree/haplotype-analysis spreadsheet, using gene order best supported by our linkage analysis to identify where recombination events had occurred.
Physical Mapping on Canine–Rodent Hybrid Cell Lines.
As described elsewhere (17), a panel of canine–rodent microcell hybrid cells were constructed by fusion of canine fibroblast donors with immortalized rodent recipient cells. Hybrid cell lines were characterized by fluorescence in situ hybridization (FISH) using dog genomic DNA probes. Three of the markers utilized in the present study (C09.250, C09.474, and C09.173), as well as two additional canine- and gene-specific sequence tagged site (STS) amplimers, were mapped onto this panel of hybrid cells to determine evidence for physical association of these markers. Primer pairs for each microsatellite or STS amplimer were used for PCR using dog, mouse, and hamster genomic DNA as templates. Cell lines from which the prcd-linked marker primers amplified an appropriately sized product were recorded as positive.
RESULTS
The set of pedigrees used in this study is illustrated in Figs. 1 and 2. Initially, a panel of approximately 100 anonymous canine-specific microsatellites (19, 36–39) was typed on a subset of the prcd-informative pedigrees. Linkage was detected between marker C09.173 (GenBank L15680 and L15681) and the prcd disease locus with 4 recombinants from 27 scored meioses [recombination fraction = 0.148, logarithm of odds (lod) score = 3.21]. This marker, C09.173, had previously been mapped to two of three canine–rodent hybrid cells—MDE6, MDE15, and MDL9 (17)—which also contained the canine homolog of the human breast cancer 1 susceptibility gene (BRCA1, GenBank U50709; refs. 17 and 44). These data enabled identification of two further anonymous microsatellites (C09.250, GenBank L15688 and L15689; and C09.474, GenBank L24352 and L24353) which mapped to the same canine–rodent hybrid cells, and a fourth (C09.2263) that was linked to C09.173 in different pedigrees. These four microsatellites were then tested on a further set of prcd-informative pedigrees, as well as on additional unrelated pedigrees to determine the linkage distances and map order of the four microsatellites (19). Analysis of these data placed prcd close to C09.173 and C09.2263, and on the opposite side of these two markers from both C09.250 and C09.474, as summarized in Table 1 and illustrated in Fig. 2A.
Table 1.
Recombination fractions and lod scores for markers flanking the prcd locus on canine chromosome 9 (CFA9)
| TK1 | GALK1 | MYL4 | 173 | 2263 | RARA | 250 | 474 | NF1 | ||
|---|---|---|---|---|---|---|---|---|---|---|
| prcd | θ | 0.000 | 0.000 | 0.033 | 0.049 | 0.051 | 0.091 | 0.156 | 0.244 | 0.320 |
| Z | 10.84 | 4.21 | 14.24 | 13.17 | 12.61 | 7.42 | 3.61 | 2.45 | 0.72 | |
| n | 36 | 14 | 70 | 61 | 61 | 37 | 47 | 41 | 25 | |
| GALK1 | θ | 0.000 | ||||||||
| Z | 4.21 | |||||||||
| MYL4 | θ | 0.039 | 0.071 | |||||||
| Z | 10.48 | 2.35 | ||||||||
| 173 | θ | 0.048 | 0.071 | 0.044 | ||||||
| Z | 12.55 | 2.65 | 15.94 | |||||||
| 2263 | θ | 0.049 | 0.071 | 0.039 | 0.000 | |||||
| Z | 12.27 | 2.65 | 16.15 | 25.29 | ||||||
| RARA | θ | 0.161 | 0.154 | 0.097 | 0.058 | 0.056 | ||||
| Z | 3.99 | 1.49 | 8.57 | 13.84 | 14.67 | |||||
| 250 | θ | 0.148 | NI | 0.121 | 0.131 | 0.139 | 0.143 | |||
| Z | 3.21 | 4.50 | 5.01 | 4.54 | 2.58 | |||||
| 474 | θ | 0.250 | 0.357 | 0.167 | 0.167 | 0.167 | 0.151 | 0.154 | ||
| Z | 1.59 | 0.25 | 3.49 | 4.42 | 4.42 | 3.84 | 1.49 | |||
| NF1 | θ | 0.368 | 0.429 | 0.277 | 0.266 | 0.276 | 0.172 | 0.00 | 0.071 | |
| Z | 0.29 | 0.06 | 0.73 | 1.48 | 1.31 | 3.27 | 4.82 | 2.65 |
Markers 173, 2263, 474, and 250 indicate canine microsatellite loci C09.173, C09.2263, C09.474, and C09.250, respectively. Data are shown in boldface where lod score exceeds 3.0. The order shown is that supported by two-point linkage analyses of data in the present study. θ, Maximum likelihood estimate of recombination fraction between loci; Z, lod score; n, number of informative meioses typed for two-point linkage analysis; and NI, no pedigrees informative.
Recognition of the physical association of these four markers, and thus prcd, to a canine chromosomal region including BRCA1 suggested that prcd was located on a canine chromosomal segment homologous to the region of human 17q where human BRCA1 maps. We therefore developed a set of PCR primers to amplify a canine-specific fragment of the γ-subunit gene for retinal cGMP phosphodiesterase (PDE6G; GenBank U49359), which we had previously cloned and sequenced (45). The human homolog of this gene is distal to BRCA1 on chromosome 17q (46). PCR amplification with these PDE6G-STS primers yielded positive results for cell lines MDE6 and MDE15 to which the loci BRCA1, C09.250, C09.474, and C09.173 had previously been mapped (17).
Thus, the canine–rodent cell lines MDE6, MDE15, and MDL9 (17) each contain all or a fragment of a canine chromosome with strong homology to human chromosome 17q24–25, and the prcd locus is physically located in this region. Because we have not yet identified any informative polymorphisms within the small (2.8-kb) PDE6G gene for these pedigrees, the linkage distance between prcd and PDE6G could not be determined.
Because of the positive result with PDE6G, we then genotyped the prcd-informative pedigrees for two restriction fragment length polymorphisms identified in a 1.5-kb amplified fragment of the canine homolog of MYL4. This locus also demonstrated linkage to prcd yielding a recombination fraction (θ) of 0.033 (lod score = 14.24).
Concurrently and independently, a set of gene-specific microsatellites were described that map to the centromeric end of CFA9 and correspond to loci on distal human chromosome arm 17q (29). We therefore typed four of these microsatellites, identified as thymidine kinase 1 (TK1), galactokinase 1 (GALK1), retinoic acid receptor α (RARA), and neurofibromin (NF1) on prcd-informative pedigrees. Two of these markers, TK1 and GALK1, cosegregated with prcd (θ = 0.000; lod scores = 10.84 and 4.21, respectively). RARA mapped to the interval bounded by markers C09.250 and C09.474 at one end and C09.173, C09.2263, and MYL4 at the other, yielding a recombination fraction of 0.091 with prcd (lod score = 7.42), which excludes this locus as a candidate for prcd. NF1 mapped further from prcd than any previous marker (θ = 0.032), but with only 25 informative individuals the lod score (0.72) associated with this linkage result was not significant. NF1 did, however, show significant linkage to either RARA (θ = 0.172; lod score = 3.27) or C09.250 (θ = 0.000; lod score = 4.82) both of which were significantly linked to prcd, thus confirming that NF1 is part of the conserved syntenic region surrounding prcd. The results of the linkage analysis are summarized in Table 1, and illustrated in Figs. 2 and 3. No obligate recombinants were observed in prcd-informative families for the interval defined by markers C09.173, C09.2263, and MYL4. Three-point analysis yielded no differences among all possible orders for these three markers together, or for any two with prcd.
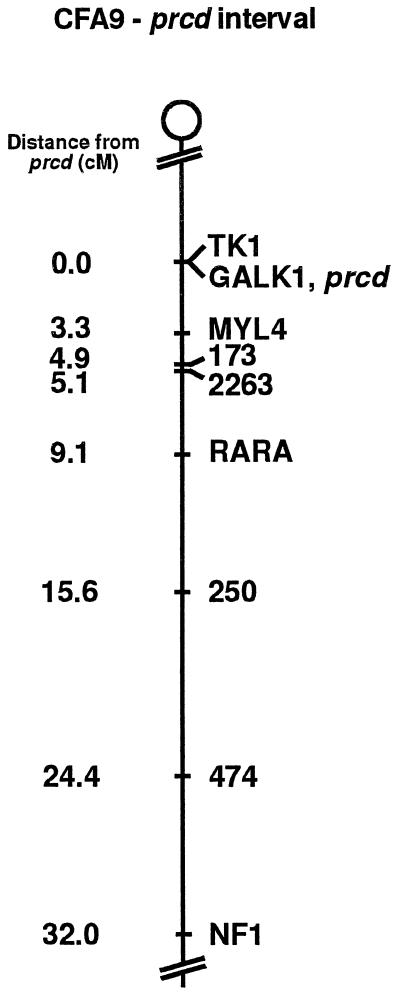
Figure 3.
Linkage map of the prcd interval on canine chromosome 9 (CFA9). The location of prcd is shown relative to that of nine loci corresponding to type I and II markers. Loci listed on a single line indicate no recombinations observed between these markers in the present study. TK1 and GALK1/prcd are on separate lines but at a single location because, although no obligate recombinations were observed in this interval, indications of possible recombinations between TK1 and prcd were detected. Order and distances (in cM, centimorgans) shown are based on two-point linkage analyses of the data in this study.
Similarly, no obligate recombinants were observed in the F2 generation of prcd-informative families for the interval defined by TK1, GALK1, and prcd. However, haplotype analysis indicates that an F1 individual, the sire of the informative litter in family 3, received a chromosome that was recombinant in the TK1-prcd-MYL4 interval from his dam (see Fig. 2B). The site of the recombination is interpretable as equally likely to be between TK1 and prcd or between prcd and MYL4. Formal linkage analysis gave a recombination distance of zero between TK1 and prcd but, if this dog is recombinant between TK1 and prcd, then the order is TK1-prcd-MYL4. The observation of two different TK1 alleles segregating in phase with prcd can also be taken to indicate recombination events taking place between TK1 and prcd in previous generations of this pedigree, lending further support to this order. Furthermore, although no recombinations were observed between GALK1 and prcd, relatively few of the typed individuals were informative for the GALK1 (CA)n repeat. Thus the supported linkage distance is broader for GALK1 (1-lod support interval = 15.2 cM) than for TK1 (1-lod support interval = 9.05 cM). The one dog indicated by haplotype analysis (Fig. 2B) as possibly recombinant between TK1 and prcd was not informative for GALK1. Three-point analysis yielded no differences among all possible orders for TK1, GALK1, and prcd.
Although, from the present data, C09.474 maps further from prcd (θ = 0.244) than does C09.250 (θ = 0.156), recombinations observed on haplotype analysis would also support placement of C09.474 between C09.250 and prcd. This order is in agreement with mapping data derived from more extensive pedigrees (19).
DISCUSSION
The linkage and cell line data presented here firmly establish that the canine prcd locus maps to the canine chromosome identified as CFA9 (12, 13, 29); previously this chromosome has also been referred to as canine chromosome 23 (14, 47). More specifically, prcd maps to the centromeric end of CFA9 in a region exhibiting conservation of synteny and gene order with distal human 17q (29, 47), mouse chromosome 11 (29, 40, 48, 49), and bovine chromosome 19 (50–52). Fig. 3 summarizes our linkage data for the centromere–NF1 region of CFA9, representing the extended prcd interval.
Linkage analysis and obligate recombinations locate prcd to the interval between TK1 and MYL4. Our data, as well as those presented elsewhere (19, 29), allow us to estimate that this distance is between 3.9 and 5.1 cM. Both TK1 and GALK1 yielded recombination fractions of 0.000 with each other and with prcd, and three-point analysis of our data does not establish a preferred order for these loci. However, haplotype analysis revealed one dog that demonstrated a possible recombination between prcd and TK1, placing TK1 on the opposite side of prcd from marker C09.173. We also observed in this dog’s family two different TK1 alleles in phase with prcd. Thus, although we did not obtain a nonzero recombination distance for the TK1--prcd interval, we do see evidence that recombinations have occurred in the extended pedigree. Similarly, no recombinants were observed between GALK1 and prcd in this study, thus placing GALK1 in the TK1–MYL4 interval. However, because relatively few individuals were informative for GALK1, our data are interpretable to support the placement of GALK1 centromeric to TK1—i.e., on the other side from prcd, which would be in better agreement with other studies (19, 29). Resolution of these issues requires additional linkage or physical mapping data.
Bardien et al. (30, 31) identified a locus (RP17) for autosomal dominant RP in two South African human kindred in the interval between markers D17S1607 and D17S1874 and close to 17q22. They initially (30) suggested three nearby genes as candidates for RP17: PDE6G, tissue inhibitor of metalloproteinases-2 (TIMP2; ref. 53), and protein kinase C α (PRKCA; refs. 54 and 55); but subsequently (31) presented evidence excluding each of these loci. RARA has also been suggested as a candidate for RP17 (31), but our data exclude it as a candidate for prcd. No genes have yet been implicated causally in RP17. Although RP17 is autosomal dominant and prcd is recessive, it is possible, perhaps likely, that these diseases arise from mutations in homologous genes. Dominant and recessive forms of RP have been identified that arise from mutations in the same gene (56, 57). The RP17 candidate region lies within the interval defined by MYL4 at 17q21–qter (ref. 40 and http://www.gdb.org), and TK1 at 17q25.2–25.3 (58), which loci also encompass the prcd interval on CFA9. As this interval forms part of the largest conserved syntenic group among mammals (40, 47–52), it seems likely that the canine homolog of RP17 lies within the corresponding interval on CFA9, and that prcd in the dog may be a true locus homolog of RP17 in humans.
Acknowledgments
The invaluable technical assistance of Gerri Antonini, Vicki Baldwin, Amanda Nickle, Susan Nitroy, Sue Pearce-Kelling, and the staff at the Retinal Disease Studies Facility, Kennett Square, PA, is gratefully acknowledged. In addition, we thank Mark Neff for contributing his data on the genotypes of these families for many anonymous microsatellite markers prior to publication. This work was supported by The Foundation Fighting Blindness, the Morris Animal Foundation/The Seeing Eye, Inc., the Cornell Center for Applied Technology, National Eye Institute Grant EY-06855, a grant from the Canine Health Foundation of the American Kennel Club to E.A.O., who is also the recipient of an American Cancer Society Junior Faculty Award, and a grant from the Canine Health Foundation of the American Kennel Club and a gift from Martin and Enid Gleich to J.R. C.S.M. is the recipient of a Wellcome International Prize Travelling Research Fellowship.
ABBREVIATIONS
- RP
retinitis pigmentosa
- ERG
electroretinogram
- STS
sequence tagged site
- lod
logarithm of odds
- cM
centimorgan
References
- 1.Little C C. The Inheritance of Coat Color in Dogs. Ithaca, NY: Comstock; 1957. [Google Scholar]
- 2.Clark R D, Stainer J R, editors. Medical and Genetic Aspects of Purebred Dogs. Edwardsville, KS: Veterinary Medicine Publishing; 1983. [Google Scholar]
- 3.Patterson D F, Haskins M E, Jezyk P F. Adv Hum Genet. 1982;12:263–339. doi: 10.1007/978-1-4615-8315-8_4. [DOI] [PubMed] [Google Scholar]
- 4.Patterson D F, Haskins M E, Jezyk P F, Giger U, Meyers-Wallen V N, Aguirre G, Fyfe J C, Wolfe J H. J Am Vet Med Assoc. 1988;193:1131–1144. [PubMed] [Google Scholar]
- 5.Bech-Nielsen S, Haskins M E, Reif J S, Brodey R S, Patterson D F, Spielman R. J Natl Cancer Inst. 1978;60:349–353. doi: 10.1093/jnci/60.2.349. [DOI] [PubMed] [Google Scholar]
- 6.Patterson D F, Pyle R L, Van Mierop L, Melbin J, Olson M. Am J Cardiol. 1974;34:187–205. doi: 10.1016/0002-9149(74)90198-2. [DOI] [PubMed] [Google Scholar]
- 7.Pyle R L, Patterson D F, Chacko S. Am Heart J. 1976;92:324–334. doi: 10.1016/s0002-8703(76)80113-5. [DOI] [PubMed] [Google Scholar]
- 8.Scott J P, Fuller J L. Genetics and Social Behavior of the Dog. Chicago: Univ. of Chicago Press; 1965. [Google Scholar]
- 9.Stockard C R. The Genetic and Endocrine Basis for Differences in Form and Behavior As Elucidated by Studies of Contrasted Pure-Line Dog Breeds and Their Hybrids, American Anatomy Memoir no. 19. Philadelphia: Wistar Inst. Anatomy and Biology; 1941. [Google Scholar]
- 10.Willis M B. Genetics of the Dog. NY: Howell Book House; 1989. [Google Scholar]
- 11.Selden J R, Moorhead P S, Oehlert M L, Patterson D F. Cytogenet Cell Genet. 1975;15:380–387. doi: 10.1159/000130537. [DOI] [PubMed] [Google Scholar]
- 12.Switonski M, Reimann N, Bosma A A, Long S, Bartnitzke S, Pienkowska A, Moreno-Milan M M, Fischer P. Chromosome Res. 1996;4:306–309. doi: 10.1007/BF02263682. [DOI] [PubMed] [Google Scholar]
- 13.Reimann N, Bartnitzke S, Bullerdiek J, Schmitz U, Rogalla P, Nolte I, Ronne M. Cytogenet Cell Genet. 1996;73:140–144. doi: 10.1159/000134326. [DOI] [PubMed] [Google Scholar]
- 14.Graphodatsky A S, Beklemisheva V R, Dolf G. Cytogenet Cell Genet. 1995;69:226–231. doi: 10.1159/000133970. [DOI] [PubMed] [Google Scholar]
- 15.Acland G M, Blanton S H, Hershfield B, Aguirre G D. Am J Med Genet. 1994;52:27–33. doi: 10.1002/ajmg.1320520106. [DOI] [PubMed] [Google Scholar]
- 16.Deschenes S M, Puck J M, Dutra A S, Somberg R L, Felsburg P J, Henthorn P S. Genomics. 1994;23:62–68. doi: 10.1006/geno.1994.1459. [DOI] [PubMed] [Google Scholar]
- 17.Langston A A, Mellersh C S, Neal C L, Ray K, Acland G M, Gibbs M, Aguirre G D, Fournier R E K, Ostrander E A. Genomics. 1997;46:317–325. doi: 10.1006/geno.1997.5009. [DOI] [PubMed] [Google Scholar]
- 18.Lingaas F, Sorensen A, Juneja R K, Johansson S, Fredholm M, Wintero A K, Sampson J, Mellersh C, Curzon A, Holmes N G, Binns M. Mamm Genome. 1997;8:218–221. doi: 10.1007/s003359900393. [DOI] [PubMed] [Google Scholar]
- 19.Mellersh C S, Langston A A, Acland G M, Fleming M A, Ray K, Weigand N A, Francisco L V, Gibbs M, Aguirre G D, Ostrander E A. Genomics. 1997;46:326–336. doi: 10.1006/geno.1997.5098. [DOI] [PubMed] [Google Scholar]
- 20.Fischer P E, Holmes N G, Dickens H F, Thomas R, Binns M M, Nacheva E P. Mamm Genome. 1996;7:37–41. doi: 10.1007/s003359900009. [DOI] [PubMed] [Google Scholar]
- 21.Acland G M, Halloran-Blanton S, Boughman J A, Aguirre G D. Am J Med Genet. 1990;35:354–359. doi: 10.1002/ajmg.1320350309. [DOI] [PubMed] [Google Scholar]
- 22.Anderson R, Maude M, Alvarez R, Acland G, Aguirre G. Exp Eye Res. 1991;52:349–355. doi: 10.1016/0014-4835(91)90100-s. [DOI] [PubMed] [Google Scholar]
- 23.Kemp C M, Jacobson S G. Exp Eye Res. 1992;54:947–956. doi: 10.1016/0014-4835(92)90159-p. [DOI] [PubMed] [Google Scholar]
- 24.Sandberg M A, Pawlyk B S, Berson E L. Invest Ophthalmol Visual Sci. 1986;27:1179–1184. [PubMed] [Google Scholar]
- 25.Aguirre G D, Acland G M. Exp Eye Res. 1988;46:663–687. doi: 10.1016/s0014-4835(88)80055-1. [DOI] [PubMed] [Google Scholar]
- 26.Ray K, Acland G M, Aguirre G D. Invest Ophthalmol Visual Sci. 1996;37:783–794. [PubMed] [Google Scholar]
- 27.Ray K, Baldwin V J, Zeiss C J, Acland G M, Aguirre G D. Curr Eye Res. 1997;16:71–77. doi: 10.1076/ceyr.16.1.71.5122. [DOI] [PubMed] [Google Scholar]
- 28.Aguirre G. In: The Second Great Basin Visual Science Symposium. Olson R J, Lasater E M, editors. Vol. 2. Salt Lake City: Univ. of Utah; 1997. pp. 6–21. [Google Scholar]
- 29.Werner P, Raducha M G, Prociuk U, Henthorn P S, Patterson D F. Genomics. 1997;42:74–82. doi: 10.1006/geno.1997.4723. [DOI] [PubMed] [Google Scholar]
- 30.Bardien S, Ebenezer N, Greenberg J, Inglehearn C F, Bartmann L, Goliath R, Beighton P, Ramesar R, Bhattacharya S S. Hum Mol Genet. 1995;4:1459–1462. doi: 10.1093/hmg/4.8.1459. [DOI] [PubMed] [Google Scholar]
- 31.Bardien S, Ramesar R, Bhattacharya S, Greenberg J. Hum Genet. 1997;101:13–17. doi: 10.1007/s004390050577. [DOI] [PubMed] [Google Scholar]
- 32.Aguirre G, O’Brien P. Invest Ophthalmol Vision Sci. 1986;27:635–655. [PubMed] [Google Scholar]
- 33.Acland G M, Aguirre G D. Exp Eye Res. 1987;44:491–521. doi: 10.1016/s0014-4835(87)80160-4. [DOI] [PubMed] [Google Scholar]
- 34.Aguirre G, Alligood J, O’Brien P, Buyukmihci N. Invest Ophthalmol Vision Sci. 1982;23:610–630. [PubMed] [Google Scholar]
- 35.Kawasaki E S. In: PCR Protocols. A Guide to Methods and Applications. Innis M A, Gelfand D H, Sninsky J J, White T J, editors. San Diego: Academic; 1990. pp. 146–152. [Google Scholar]
- 36.Francisco L V, Langston A A, Mellersh C S, Neal C L, Ostrander E A. Mamm Genome. 1996;7:359–362. doi: 10.1007/s003359900104. [DOI] [PubMed] [Google Scholar]
- 37.Ostrander E A, Sprague G F, Jr, Rine J. Genomics. 1993;16:207–213. doi: 10.1006/geno.1993.1160. [DOI] [PubMed] [Google Scholar]
- 38.Ostrander E A, Mapa F A, Yee M, Rine J. Mamm Genome. 1995;6:192–195. doi: 10.1007/BF00293011. [DOI] [PubMed] [Google Scholar]
- 39.Ostrander E A, Jong P M, Rine J, Duyk G. Proc Natl Acad Sci USA. 1992;89:3419–3423. doi: 10.1073/pnas.89.8.3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Seharaseyon J, Bober E, Hsieh C L, Fodor W L, Francke U, Arnold H H, Vanin E F. Genomics. 1990;7:289–293. doi: 10.1016/0888-7543(90)90554-8. [DOI] [PubMed] [Google Scholar]
- 41.Lathrop G M, Lalouel J M. Am J Hum Genet. 1984;36:460–465. [PMC free article] [PubMed] [Google Scholar]
- 42.Terwilliger J D, Ott J. Handbook of Human Genetic Linkage. Baltimore: Johns Hopkins Univ. Press; 1994. [Google Scholar]
- 43.Ott J. Analysis of Human Genetic Linkage. Baltimore: Johns Hopkins Univ. Press; 1991. [Google Scholar]
- 44.Szabo C I, Wagner L A, Francisco L V, Roach J C, Argonza R, King M-C, Ostrander E A. Hum Mol Genet. 1996;5:1289–1298. doi: 10.1093/hmg/5.9.1289. [DOI] [PubMed] [Google Scholar]
- 45.Wang W, Acland G M, Aguirre G D, Ray K. Gene. 1996;181:1–5. doi: 10.1016/s0378-1119(96)00319-8. [DOI] [PubMed] [Google Scholar]
- 46.Hall J M, Lee M K, Newman B, Morrow J E, Anderson L A, Huey B, King M-C. Science. 1990;250:1684–1689. doi: 10.1126/science.2270482. [DOI] [PubMed] [Google Scholar]
- 47.Park J P. Cytogenet Cell Genet. 1996;74:133–137. doi: 10.1159/000134400. [DOI] [PubMed] [Google Scholar]
- 48.Andersson L, Archibald A, Ashburner M, Audun S, Barendse W, Bitgood J, Bottema C, Broad T, Brown S, Burt D, Charlier C. Mamm Genome. 1996;7:717–734. doi: 10.1007/s003359900222. [DOI] [PubMed] [Google Scholar]
- 49.Eppig J T. Curr Opin Genet Dev. 1996;6:723–730. doi: 10.1016/s0959-437x(96)80027-x. [DOI] [PubMed] [Google Scholar]
- 50.Solinas-Toldo S, Lengauer C, Fries R. Genomics. 1995;27:489–496. doi: 10.1006/geno.1995.1081. [DOI] [PubMed] [Google Scholar]
- 51.Yang Y P, Womack J E. Genomics. 1995;27:293–297. doi: 10.1006/geno.1995.1045. [DOI] [PubMed] [Google Scholar]
- 52.Yang Y P, Womack J E. Mamm Genome. 1997;8:262–266. doi: 10.1007/s003359900406. [DOI] [PubMed] [Google Scholar]
- 53.Hammani K, Blakis A, Morsette D, Bowcock A M, Schmutte C, Henriet P, DeClerck Y A. J Biol Chem. 1996;271:25498–25505. doi: 10.1074/jbc.271.41.25498. [DOI] [PubMed] [Google Scholar]
- 54.Finkenzeller G, Marme D, Hug H. Nucleic Acids Res. 1990;18:2183. doi: 10.1093/nar/18.8.2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Summar M L, Phillips J A, 3d, Krishnamani M R, Keefer J, Trofatter J, Schwartz R C, Tsipouras P, Willard H, Ullrich A. Genomics. 1989;5:163–165. doi: 10.1016/0888-7543(89)90104-3. [DOI] [PubMed] [Google Scholar]
- 56.Danciger M, Blaney J, Gao Y-Q, Zhao D-Y, Heckenlively J R, Jacobson S G, Farber D B. Genomics. 1995;30:1–7. doi: 10.1006/geno.1995.0001. [DOI] [PubMed] [Google Scholar]
- 57.Rosenfeld P J, Cowley G S, McGee T L, Sandberg M A, Berson E L, Dryja T P. Nat Genet. 1992;1:209–213. doi: 10.1038/ng0692-209. [DOI] [PubMed] [Google Scholar]
- 58.Kuo W-L, Hirschhorn R, Huie M L, Hirschhorn K. Hum Genet. 1996;97:404–406. doi: 10.1007/BF02185782. [DOI] [PubMed] [Google Scholar]