Abstract
Malate plays a central role in plant metabolism. It is an intermediate in the Krebs and glyoxylate cycles, it is the store for CO2 in C4 and crassulacean acid metabolism plants, it protects plants from aluminum toxicity, it is essential for maintaining the osmotic pressure and charge balance, and it is therefore involved in regulation of stomatal aperture. To fulfil many of these roles, malate has to be accumulated within the large central vacuole. Many unsuccessful efforts have been made in the past to identify the vacuolar malate transporter; here, we describe the identification of the vacuolar malate transporter [A. thaliana tonoplast dicarboxylate transporter (AttDT)]. This transporter exhibits highest sequence similarity to the human sodium/dicarboxylate cotransporter. Independent T-DNA [portion of the Ti (tumor-inducing) plasmid that is transferred to plant cells] Arabidopsis mutants exhibit substantially reduced levels of leaf malate, but respire exogenously applied [14C]malate faster than the WT. An AttDT-GFP fusion protein was localized to vacuole. Vacuoles isolated from Arabidopsis WT leaves exhibited carbonylcyanide m-chlorophenylhydrazone and citrate inhibitable malate transport, which was not stimulated by sodium. Vacuoles isolated from mutant plants import [14C]-malate at strongly reduced rates, confirming that this protein is the vacuolar malate transporter.
In all organisms, malate and citrate fulfil a wide range of important functions, the most universal one being their role in the Krebs cycle. Nearly all eukaryotic and prokaryotic cells possess transport proteins directing import of these metabolites (1, 2). In plants, so far, dicarboxylate transporters have been identified only in mitochondria and chloroplasts (3, 4). In contrast, the existence of a vacuolar malate/citrate transporter has been described only at the functional level by transport studies using either intact vacuoles or vacuolar membrane vesicles, by flux or patch clamp analysis (5–7). Flux studies revealed a malate transporter exhibiting apparent KM values of 1–4 mM, and electrophysiological studies supported the existence of a malate channel protein. Malate storage within the vacuole allows the plant to accumulate this metabolite to very high concentrations (up to >300 mM) and thus maintain constant the cytosolic concentration (8, 9).
In our efforts to identify the vacuolar dicarboxylate transporter on the molecular level, we searched for a putative candidate gene. In mammalian cells, dicarboxylate import is mediated by transport proteins that are energized by a sodium gradient (1). The human sodium/dicarboxylate cotransporter (HsNaDC-1), the first of various human isoforms to be identified on the molecular level, resides in the plasma membrane of the renal proximal kidney tubule and is the major importer for the retrieval of di- and tricarboxylates from the primary urine (10, 11). Such sodium/dicarboxylate cotransporters are not restricted to kidney cells; they generally reside in plasma membranes, comprise ≈600 aa, and exhibit 11–12 predicted transmembrane domains (1).
We discovered a single copy gene in Arabidopsis encoding a protein with substantial sequence homology to the sodium/ dicarboxylate cotransporter located in human kidneys. Because close homologs of kidney aquaporins reside both at the plant plasma membrane and at the vacuolar membrane, we were interested to investigate the location and the function of the plant protein homolog to the human kidney sodium/dicarboxylate cotransporter. Physiological, biochemical, and molecular analyses on WT and knockout plants indicate that this Arabidopsis transporter does not reside in the plasma membrane but represents the carrier protein responsible for malate uptake into the vacuole.
Materials and Methods
Plant Cultivation and Isolation of the Full-Length AttDT cDNA Clone. WT and transgenic Arabidopsis thaliana (ecotype Wassilewskija) were cultivated in a growth chamber in soil at 20°C with a light intensity of 150 μE·m-2·s-1. Light was present for 10 h per day. For Northern blot analysis, WT plants were alternatively grown for 14 days on agar medium (12), supplemented with increasing malate levels.
The full-length AttDT cDNA was cloned from an A. thaliana plasmid cDNA library kindly provided by M. Minet (Centre National de la Recherche Scientifique, Gif-sur-Yvette, France). The library was screened with a homologous AttDT cDNA probe according to standard protocols (13). The homologous cDNA was 32P-radiolabeled by random priming (Ready-To-Go DNA labeling beads, Amersham Pharmacia Biosciences). Prehybridization and hybridization were performed at 42°C in Church buffer medium (13). The required sequence information to generate the AttDT cDNA probe was taken from the Arabidopsis genome project (www.tigr.org).
Extraction of Total RNA and RNA Gel-Blot Hybridization. Total RNA was isolated from frozen tissue samples by using the Purescript RNA extraction kit (Gentra Systems) according to the manufacturer's instructions. RNA samples (10 μg) were denatured and separated on a 1.2% agarose/2.5% formaldehyde gel. Ethidium bromide was included in the loading buffer medium to confirm equal sample loading.
Isolated RNA were blotted onto a nylon membrane (Nytran-Plus, Schleicher & Schüll, Dassel, Germany), and AttDT cDNA was 32P-labeled by random priming (Ready-To-Go DNA labeling beads, Amersham Pharmacia). Prehybridization and hybridization were performed at 65°C in Church buffer medium. Final blots were exposed to Kodak MS x-ray film at -70°C, or visualized by a PhosphorImager (Packard).
AttDT-GFP Fusion and Fluorescence Microscopy. To construct the AttDT-GFP fusion protein, we amplified (by means of PCR) the entire AttDT cDNA by using the Pfu-DNA polymerase. The obtained DNA product of AttDT was cleaved with XbaI and XhoI and inserted in-frame in the vector GFP2 (14), leading to the final AttDT-GFP construct. A plasmid containing the tomato sucrose transporter LeSUT1-GFP fusion protein construct was kindly provided by M. Tegeder and W. Frommer (Universität Tübingen, Tübingen, Germany). Protoplasts, either isolated from sterile grown tobacco (Nicotiana tabacum cv. W38) or from soil grown Arabidopsis plants, were transfected with column-purified plasmid DNA (30 μg per 0.5 × 106 cells). After 1 day incubation at 24°C in the dark, protoplasts in Petri dishes were checked for the presence of green fluorescing cells by using a laser scanning confocal microscope (TCS 4D, Leica, Bensheim, Germany). GFP was excited at 488 nm, and the emission was detected by a photomultiplier through a 530/30-nm band pass filter. Vacuoles from transfected Arabidopsis protoplasts were enriched (15) and checked for green fluorescence (at 470 nm) by using a standard Zeiss fluorescence microscope (Axioskop).
AttDT Mutant Screen. The Arabidopsis knockout mutant lines AttDT::tDNA-1 and AttDT:tDNA-2 were identified by screening a library of 64,000 individual tDNA lines (Arabidopsis Knockout Facility, University of Wisconsin Biotechnology Center) by PCR for T-DNA [portion of the Ti (tumor-inducing) plasmid that is transferred to plant cells] insertion within the gene region of AttDT. Genomic DNA was isolated from plant tissues by standard methods. Seeds from positive pools of T-DNA insertion lines were grown, and individual plants were screened by PCR as described below. The following gene- and T-DNA-specific primers were used: AttDT-specific primers AttDT1 (5′-GGTCTTACACCGAATATTTTCGTCATATG-3′) and AttDT2 (5′-AATCAATTGTGTACATGGTCTTGTGCTTG-3′), left border primer JL202 (5′-CATTTTATAATAACGCTGCGGACATCTAC-3′).
Determination of Leaf Malate Content. Leaf material was prepared with a cork borer (5-mm diameter) from WT or AttDT::tDNA-1 plants at the indicated time points after onset of illumination. The material was kept under continuous illumination, then immediately weighed, subsequently snap frozen in liquid nitrogen, and extracted in a chloroform/methanol-buffer medium. Malate in the sample was quantified photometrically according to a routine protocol on a Sigma-Eppendorf dual wavelength photometer (16).
Quantification of Respiratory Use of [14C]Malate. For analysis of the [14C]malate-driven respiratory activity, we prepared leaf discs (5-mm diameter), 3 h after onset of illumination, from WT and knockout mutants (grown for ≈3–4 wk in the growth chamber) in 2-ml reaction tubes with 100 μl of 100 μM [14C]malate [at a specific activity of 0.4 μCi/μl(1Ci = 37 GBq)] and 20 mM NaCl. On the inside at the top of this tube, a small 0.5-ml reaction vessel containing 50 μl of 1M KOH was fixed with grease to allow 14CO2 absorption. The leaf discs were allowed to float on the solution, and incubation continued for 6 or 24 h in the dark. The reaction was stopped by adding 100 μl of 2 M HCl with a syringe through the closed lid. The hole in the lid was sealed with grease, and, after subsequent incubation for 10 h, the released radioactively labeled CO2 was quantified in a scintillation counter.
Quantification of [14C]Malate Uptake into Isolated Arabidopsis Vacuoles. For isolation of intact Arabidopsis vacuoles, 3- to 4-wk-old WT and knockout mutant plants grown in soil were used. Digestion of cells and isolation of vacuoles were performed essentially as described (15). Transport studies of [14C]malate into isolated vacuoles were performed as described (6) by using the silicone-oil centrifugation technique. The uptake medium was free of sodium and contained potassium gluconate instead of potassium chloride. To energize isolated vacuoles, ATP was present at a concentration of 1 mM.
Results and Discussion
Up to now, all attempts have failed to identify the molecular nature of the plant vacuolar dicarbonic acid transporter (17). A biochemical approach to isolate the corresponding protein is extremely hampered by the low number of tonoplast membranes usually enriched from plant tissues (18, 19).
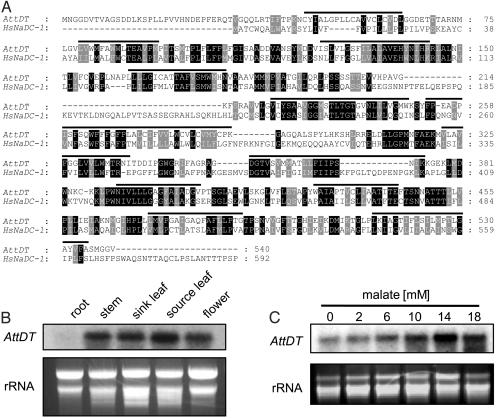
Interestingly, the A. thaliana genome contains, on chromosome 5, a single copy of an ORF encoding a highly hydrophobic protein that exhibits highest sequence similarity to HsNaDC-1 (65.6% similarity, 38.4% identity, Fig. 1A). The putative protein comprises 540 aa in length, is predicted to contain 12 transmembrane domains, and possesses a calculated molecular mass of 59.4 kDa (Fig. 1 A). The N-terminal domain, which extends the Arabidopsis protein in comparison with the human homolog, does not exhibit signatures typically required for mitochondrial or chloroplastic targeting (Fig. 1 A). The Arabidopsis protein [A. thaliana tonoplast dicarboxylate transporter (AttDT)] does not exhibit considerable amino acid sequence homology to the dicarbonic acid transport proteins recently identified in plant mitochondria and chloroplast, respectively (3, 4).
Fig. 1.
Alignment of amino acid sequences of AttDT and HsNaDC-1 (A), tissue specificity of AttDT transcript accumulation (B), and effect of malate on leaf AttDT mRNA levels (C). (A) AttDT, EMBL accession no. AJ223445; HsNaDC-1, EMBL accession no. NP003975. The alignment was calculated with the “pileup” program in gcg (22). Bars represent the predicted transmembrane domains (23). (B) Tissues were collected from Arabidopsis plants grown on soil under ambient conditions. (C) WT plants were grown for 14 days on agar medium supplemented with increasing malate levels. The experiments were carried out three times with similar results.
A Northern blot analysis confirmed that the AttDT gene is expressed. The corresponding transcript predominantly accumulates in fully developed source leaves, followed by sink leaves and stem and flower tissue, but it is hardly expressed in roots (Fig. 1B). This expression pattern indicates that AttDT has a major function in tissues harboring photosynthetically active cells.
To obtain a first indication of a physiological function of AttDT, Arabidopsis seedlings were grown for 14 days on agar medium supplemented with increasing malate concentrations, and the AttDT transcript levels were determined. Malate caused a strong increase in the AttDT mRNA in leaves, a 14-mM concentration being most effective (Fig. 1C). This observation gave an experimental indication of a physiological relationship between AttDT and dicarboxylic acids.
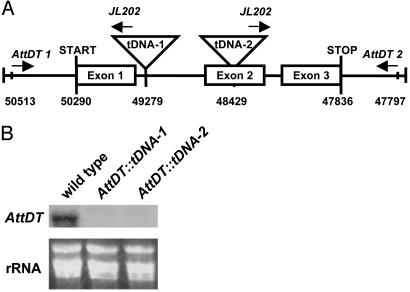
Two independent AttDT-t-DNA knockout lines were identified in Arabidopsis and used to gain insight into the function of AttDT in leaves. Knockout lines AttDT::tDNA-1 and AttDT::tDNA-2 harbor a T-DNA insertion at positions 49279 and 48429, respectively, of chromosome 5 (Fig. 2A). The combination of the AttDT-specific primers AttDT1 and AttDT2 allowed amplification of a corresponding PCR product (2,714 bp) on genomic DNA from WT, but not on DNA from homozygous AttDT knockout plants from lines 1 and 2 (data not shown). A 1,360-bp junction sequence was amplified in reactions containing the T-DNA left border primers JL202 and AttDT1 on DNA from homozygous AttDT knockout line 1, but not on DNA extracted from WT plants (data not shown). Similarly, we screened the second AttDT knockout mutant line 2. The gene-specific primer AttDT2 and the left border primer JL202 led to the amplification of a PCR product of 759 bp from genomic knockout DNA (data not shown), but not in DNA from WT plant. The amplified PCR fragments were sequenced to confirm the presence of an AttDT sequence and to determine the T-DNA insertion site.
Fig. 2.
Structure of the two independent T-DNA insertions into the AttDT gene and Northern blot analysis of AttDT mRNA accumulation in knockout leaves. (A) Structure of the T-DNA insertions into the AttDT gene. (B) Northern blot analysis of AttDT-mRNA accumulation in knockout lines. Leaf material was collected from plants grown in soil under ambient conditions.
PCR analysis of genomic DNA revealed that, in both cases, homozygous T-DNA lines had been generated by backcrossing of the heterozygous lines (see above), and both homozygous knockout lines lack the AttDT transcript as revealed by Northern blot analysis (Fig. 2B) and RT-PCR (data not shown).
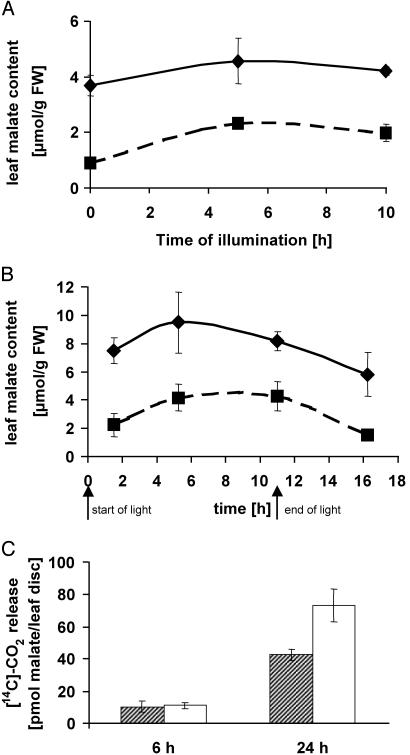
We quantified the total malate levels in leaves from WT and knockout plants. These plants were grown under ambient conditions and were phenotypically indistinguishable. However, the malate levels in WT leaves were up to four times higher compared with leaves of the knockout lines (Fig. 3A). At the beginning of the light period, the malate content in leaves was ≈3.7 and 0.9 μmol of malate per gfw (gfw, gram fresh weight) in WT and AttDT::tDNA-1 plants, respectively (Fig. 3A). A similar difference was observed during the entire day. At the end of the light period, leaf malate content was slightly increased, i.e., 4.2 μmol/gfw in WT and 1.9 μmol of malate per gfw in AttDT::tDNA-1 plants (Fig. 3A). For AttDT::tDNA-2, essentially similar results were obtained (data not shown).
Fig. 3.
Malate content in leaves from WT and AttDT::t-DNA-1 plants and respiratory degradation of [14C]malate applied to leaf discs from WT and AttDT::t-DNA-1 plants. (A) Malate content in leaves from WT and AttDT::t-DNA-1 plants. Leaf material was prepared from WT (solid line) or AttDT::tDNA-1 (dashed line) plants. The data presented are the mean (±SE) of three independent experiments. (B) Effect of sodium chloride application on leaf malate content in WT and AttDT::t-DNA-1 plants. Before extraction, plants were watered for 14 days with 50 mM NaCl. Solid line represents malate in WT leaves; dashed line represents malate in AttDT::tDNA-1 plants. The data are the mean (±SE) of four independent experiments. (C) Respiratory degradation of [14C]malate applied to leaf discs from WT and AttDT::tDNA-1 plants. Leaf discs (5-mm diameter) were prepared from intact plants, grown on soil under ambient conditions, with a cork borer 3 h after onset of illumination. For details, see Materials and Methods. The data represent the mean (±SE) of three independent experiments. Hatched bars, leaf discs from WT plants; open bars, leaf discs from AttDT::t-DNA-1 plants.
Despite the fact that WT and AttDT knockout lines exhibit essentially the same morphological phenotype under standardized growing conditions, they showed a markedly different biochemical phenotype when grown on high sodium chloride concentrations. The application of 50 mM NaCl for 2 wk corresponds to significantly increased leaf malate levels (Fig. 3B). During the day in WT plants, the malate concentration of ≈9 μmol of malate per gfw is twice as high as in water controls (Fig. 3 A and B) and significantly higher than in corresponding AttDT::tDNA-1 plants (Fig. 3B). These results indicate that leaf malate levels in Arabidopsis increase on salt stress, probably to compensate for the charge difference between sodium and chloride taken up by the plant. In addition, these results further reveal a biochemical difference between WT and AttDT knockout plants.
In leaves from various plant species, the vacuole fills ≈75% of the mesophyll cell volume (8, 20). Furthermore, the cytosolic malate concentration was found to be constant (9) at a concentration of ≈2 mM. Therefore, excess malate is transported to and stored within the central vacuole (8). Thus, the observation of large differences in the malate content in leaves of WT and knockout plants strongly suggests that the absence of AttDT results in a substantially decreased malate content in the vacuole.
To obtain information on the subcellular location of AttDT, we fed radioactively labeled malate to leaf discs prepared from either WT or AttDT::tDNA-1 plants. Within the first 6 h of incubation, leaf discs from AttDT::tDNA-1 plants respired exogenously applied [14C]malate only slightly faster than corresponding WT leaves (Fig. 3C). However, within 24 h of incubation, discs from AttDT::tDNA-1 leaves respired [14C]malate at a rate of 74 pmol/mm2, whereas the WT tissue respired only 42.5 pmol of malate per mm2 (Fig. 3C). Again, leaf discs prepared from the second knockout line, AttDT::tDNA-2, behaved similarly (data not shown). These data suggest that AttDT is not an import protein located in the plasma membrane because the absence of this carrier protein does obviously not prevent the entry of labeled malate into the cells of knockout plants. On the other hand, to our knowledge facilitated export of malate from mesophyll cells into the apoplast has never been reported. Therefore, the increased respiratory use of exogenously provided malate by the knockout lines is in accordance with the presumed tonoplastic localization of AttDT. In knockout cells, it can be assumed that the vacuolar compartmentation of newly imported malate is impaired, resulting in higher cytosolic [14C]malate levels compared with the situation in WT cells. This change results in an increased mitochondrial use of malate in the knockout cells. Taken together, these results further indicate that AttDT is located in the vacuolar membrane rather than (like the mammalian homolog) in the plasma membrane.
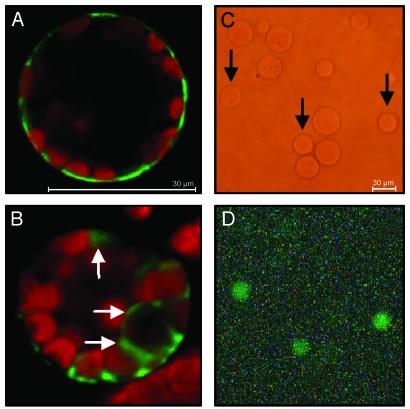
To provide further, independent evidence for a vacuolar localization of AttDT, we transfected tobacco protoplasts with a plasmid construct directing the synthesis of a AttDT-GFP fusion protein and analyzed its localization. As an internal control, we also analyzed the localization of a GFP protein carrying at its N terminus the sucrose transporter from tomato (LeSUT1), which is known to reside in the plasma membrane (21). Expression of the LeSUT1-GFP protein in tobacco protoplasts results in green fluorescence exclusively in the plasma membrane (Fig. 4A). In contrast, expression of AttDT-GFP fusion protein results in green fluorescence of internal membrane structures (Fig. 4B), especially visible between the chloroplasts and around the large nucleus (Fig. 4B), clearly indicating that the AttDT-GFP fusion protein is present in the tonoplast. The tonoplastic localization of the AttDT protein has finally been demonstrated by analysis of the green fluorescence of intact vacuoles enriched from Arabidopsis protoplasts transfected with the plasmid allowing AttDT-GFP expression (Fig. 4 C and D). As shown in Fig. 4D, several of the single vacuoles exhibited strong green fluorescence, and the number of fluorescing vacuoles reflects the transformation efficiency. AtDT-GFP always accumulated in the major and not in small vacuoles (Fig. 4D).
Fig. 4.
Subcellular localization of the AttDT-GFP fusion protein in transfected protoplasts. (A) Localization of the tomato sucrose transporter LeSUT1-GFP fusion protein in tobacco protoplasts. (B) Localizations of the AttDT-GFP fusion protein in tobacco protoplasts. (C) Vacuoles enriched from Arabidopsis protoplasts expressing the AttDT-GFP construct and analyzed under standard white light. (D) The same vacuoles as in C analyzed with light exciting GFP (at 470 nm). White arrows indicate areas of highest green fluorescence; black arrows highlight the position of single vacuoles exhibiting green fluorescence.
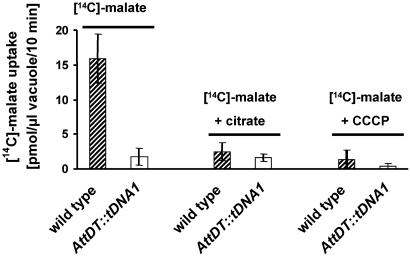
For direct and unequivocal confirmation that AttDT is indeed the vacuolar malate transporter, we isolated intact vacuoles from WT and knockout leaves and performed flux analyses by using [14C]malate. Vacuoles from WT leaves imported malate at a rate of 15.9 pmol of malate per μl of vacuole per 10 min (Fig. 5), whereas the rate for vacuoles from the knockout AttDT::tDNA-1 was 8.5 times smaller (1.8 pmol of malate per μl of vacuole per 10 min, Fig. 5). This result provides the proof that AttDT is indeed the vacuolar malate transporter. It is tempting to speculate that the low residual malate transport observed in the vacuoles of the knockout plants corresponds to the activity of the malate channel reported in the literature (7).
Fig. 5.
[14C]Malate uptake into vacuoles isolated from WT or AttDT::tDNA-1 plants. [14C]Malate was given at a concentration of 70 μM. If indicated, citrate was present at a concentration of 350 μM; if indicated, the uncoupler carbonylcyanide m-chlorophenylhydrazone (CCCP) was present at a concentration of 20 μM. Uptake was allowed for 10 min and was linear within this time. The given data represent the mean (±SE) of three independent experiments. Hatched bars, leaf discs from WT plants; open bars, leaf discs from AttDT::tDNA-1 plants.
We performed inhibitor studies to further underline that the Arabidopsis protein AttDT corresponds to the vacuolar malate carrier previously characterized by transport studies on intact vacuoles. The import of malate into WT vacuoles is strongly inhibited by the addition of citrate. Citrate in 5-fold excess (350 μM) decreased [14C]malate uptake by WT vacuoles to ≈16% of the control (Fig. 5). It has been reported (6) that citrate is a competitive inhibitor of the vacuolar malate transporter and exhibits a higher affinity than malate. In contrast, the residual malate transport activity observed for the knockout plants was not inhibited by citrate (Fig. 5). Similarly, uptake of malate by WT vacuoles is strongly reduced by the uncoupler carbonylcyanide m-chlorophenylhydrazone, but less in vacuoles isolated from the knockout plants (Fig. 5). In contrast to the dependence of the human dicarboxylate carrier on sodium, no stimulating effect of sodium could be observed (not shown).
Conclusions
A large number of studies have shown that transport of malate into the vacuole and its subsequent storage are essential in, e.g., stomatal opening and crassulacean acid metabolism metabolism. In the present study, we have identified the gene encoding the vacuolar malate transporter in Arabidopsis. Plants knocked out in the AttDT gene lack the vacuolar malate transport activity, they accumulate less malate in the leaves, and respire exogenous given malate at an increased rate. Thus, in contrast to the homolog in animal cells, the plant protein resides in the subcellular tonoplast membrane. In addition, malate transport catalyzed by AttDT is not energized by sodium. It will be interesting to extend this study to the identification of the corresponding malate transporter in crassulacean acid metabolism plants and other species exhibiting a highly active cellular malate metabolism.
Acknowledgments
This work is dedicated to the late Dr. Rafael Ratajczak. We thank Drs. Christian Lohr and Torsten Möhlmann (University of Kaiserslautern) for expert help during the confocal analysis, Prof. N. Amrhein (Eidgenössische Technische Hochschule, Zürich) for critical reading of the manuscript, and Esther Vogt (University of Zürich) for help in performing some uptake experiments. The work in the laboratory from H.E.N. was supported by the Deutsche Forschungsgemeinschaft, Graduiertenkolleg 845/1. E.M. is supported by the Bundesamt für Bildung und Wissenschaft within the project Novel Ion Channels in Plants (BBW 01.0598; EU HPRN-CT-00245).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: AttDT, Arabidopsis thaliana tonoplast dicarboxylate transporter; HsNaDC-1, human sodium/dicarboxylate cotransporter; T-DNA, portion of the Ti (tumor-inducing) plasmid that is transferred to plant cells; gfw, gram fresh weight.
Data deposition: The sequence for AttDT reported in this paper has been deposited in the EMBL database (accession no. AJ223445).
References
- 1.Pajor, A. M. (1999) Annu. Rev. Physiol. 61, 663-682. [DOI] [PubMed] [Google Scholar]
- 2.Jung, H. (2001) Biochim. Biophys. Acta 1505, 131-143. [DOI] [PubMed] [Google Scholar]
- 3.Catoni, E., Schwab, R., Hilpert, M., Desimone, M., Schwacke, R., Flügge, U. I., Schumacher, K. & Frommer, W. B. (2003) FEBS Lett. 534, 87-92. [DOI] [PubMed] [Google Scholar]
- 4.Weber, A., Menzlaff, E., Arbinger, B., Gutensohn, M., Eckerskorn, C. & Flügge, U. I. (1995) Biochemistry 34, 2621-2627. [DOI] [PubMed] [Google Scholar]
- 5.Buser, C. & Matile, P. (1977) Z. Pflanzenphysiol. 82, 462-466. [Google Scholar]
- 6.Rentsch, D. & Martinoia, E. (1991) Planta 184, 532-537. [DOI] [PubMed] [Google Scholar]
- 7.Pantoja, O. & Smith, J. A. C. (2002) J. Membr. Biol. 186, 31-42. [DOI] [PubMed] [Google Scholar]
- 8.Winter, H., Robinson, D. G. & Heldt, H. W. (1994) Planta 193, 530-535. [Google Scholar]
- 9.Gerhardt, R., Stitt, M. N. & Heldt, H. W. (1987) Plant Physiol. 83, 339-407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pajor, A. M. (1996) Am. J. Physiol. 270, 642-648. [Google Scholar]
- 11.Wright, E. M. (1985) Annu. Rev. Physiol. 47, 127-141. [DOI] [PubMed] [Google Scholar]
- 12.Weigel, D. & Glazebrook, J. (2002) Arabidopsis: A Laboratory Manual (Cold Spring Harbor Lab. Press, Plainview, NY).
- 13.Sambrook, J., Fritsch, E. F. & Maniatis, T. (1989) Molecular Cloning: A Laboratory Manual (Cold Spring Harbor Lab. Press, Plainview, NY).
- 14.Kost, B., Spielhofer, P. & Chua, N.-H. (1998) Plant J. 16, 393-401. [DOI] [PubMed] [Google Scholar]
- 15.Frangne, N., Eggmann, T., Koblischke, C., Weissenböck, G., Martinoia, E. & Klein, M. (2002) Plant Physiol. 128, 726-733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Passonneau, L. V. & Lowry, O. H. (1993) Enzymatic Analysis: A Practical Approach (Humana, Totowa, NJ).
- 17.Martinoia, E., Massoneau, A. & Frangne, N. (2000) Plant Cell Physiol. 41, 1175-1181. [DOI] [PubMed] [Google Scholar]
- 18.Martinoia, E., Vogt, E., Rentsch, D. & Amrhein, N. (1991) Biochim. Biophys. Acta 1062, 271-278. [DOI] [PubMed] [Google Scholar]
- 19.Steiger, S., Pfeifer, T., Ratajczak, R., Martinoia, E. & Lüttge, U. (1997) J. Plant Physiol. 151, 137-141. [Google Scholar]
- 20.Winter, H., Robinson, D. G. & Heldt, H. W. (1993) Planta 191, 180-190. [Google Scholar]
- 21.Ward, E. R., Kuhn, C., Tegeder, M. & Frommer, W. B. (1998) Int. Rev. Cytol. 178, 41-71. [DOI] [PubMed] [Google Scholar]
- 22.Devereux, J., Haberli, P. & Smithies, O. (1984) Nucleic Acids Res. 12, 387-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kyte, J. K. & Doolitle, R. F. (1982) J. Mol. Biol. 157, 105-132. [DOI] [PubMed] [Google Scholar]