Abstract
The B subunit of Escherichia coli heat labile enterotoxin (LT-B) is a potent oral immunogen with potential for use as a vaccine, a carrier molecule to deliver antigens to gut-associated lymphoid tissues, and possibly an adjuvant to make coadministered vaccines more effective. LT-B produced in plants was shown to be functional and immunogenic in animals and humans. In this work, we show that maize-derived LT-B is strongly associated with starch granules in endosperm. Using immunogold labeling/electron microscopy, cell fractionation, and protein analysis techniques, we observed that LT-B protein could be detected both internally and externally in starch granules. This strong association confers an effective copurification of the antigen with the starch fraction of maize kernels, thermostability desirable in maize processing, and resistance to peptic degradation in simulated gastric fluid digests, an important attribute for an orally delivered antigen.
Keywords: LT-B, starch localization
The Escherichia coli heat labile enterotoxin B subunit (LT-B), a potent oral immunogen, has been used as a model antigen to demonstrate the feasibility of producing an effective oral vaccine in transgenic plants (1–3). Orally administered LT-B has been shown to elicit strong mucosal and serum antibody responses. LT-B could be used as an adjuvant, stimulating immune responses against coadministered antigens (4). In addition, LT-B's receptor-binding capacity in the gut makes it an ideal carrier molecule for the delivery of antigenic epitopes to the gut's mucosal system (5).
The likelihood of protein degradation in the stomach and uncertainty about the effectiveness of orally delivered antigens has raised concerns about the oral vaccination approach. The delivery of an oral vaccine in transgenic plant tissue could protect it from the low pH and digestive enzymes of the stomach as well as prolong the exposure of antigen to the gut's immune system. The subcellular location of any novel protein expressed in plants is also important for its accumulation, folding, and assembly, and, depending on its intended use, may have an effect on their functionality (6).
Proteins are commonly targeted to extracytosolic compartments in both eukaryotic and prokaryotic cells, which occurs by a variety of mechanisms. Translocation mechanisms usually involve the synthesis of a protein with a specific targeting signal and the recognition of this signal by the appropriate translocation machinery (7). Numerous bacterial and viral proteins have been produced in transgenic plants. Some proteins were transported into chloroplasts when they were fused to chloroplast transit peptide (8, 9). It is generally assumed that the subcellular destinations for those heterologous proteins produced in transgenic plants will be determined by the subcellular targeting information contained in the protein, and an appropriate targeting signal is chosen based on the preferred destination for the protein. In this study, we determined the subcellular destination of the endosperm-expressed bacterial protein, LT-B, in transgenic maize kernels, when it was produced with either its native bacterial signal peptide or a plant signal peptide. In addition, we analyzed the practical significance of the observed LT-B distribution in maize endosperm.
Materials and Methods
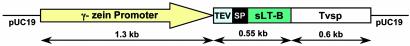
DNA Constructs and Plant Transformation. The LT-B gene cassettes were regulated by the maize endosperm-specific 27-kDa γ-zein promoter (10), tobacco etch virus translational enhancer leader sequence (11), and soybean vegetative storage protein terminator (12), as shown in Fig. 1. The synthetic LT-B gene (sLT-B) coding sequence was optimized for a compromise between maize and potato codon usage (1). Construct P77 contains the native LT-B bacterial signal peptide (1), and construct P81 contains the maize 27-kDa γ-zein signal peptide (10). Transformation of maize plants with constructs P77 and P81 (Fig. 1) was achieved by using the biolistic-gun method (13), and transgenic plants were grown to maturity in the greenhouse.
Fig. 1.
Schematic diagram of constructs P77 and P81 used for maize transformation to generate LT-B-expressing transgenic plants. Both constructs are in a pUC19 vector and contain the 27-kDa maize γ-zein promoter, tobacco etch virus (TEV) translational enhancer leader sequence, sLT-B, and soybean vegetative storage protein terminator (Tvsp). SP, signal peptide. P77, sLT-B fused to the bacterial signal peptide from LT-B; P81, sLT-B fused to signal peptide from the maize 27 kDa γ-zein protein.
Fixation of Tissue for Immunolocalization. Immature (23 d postpollination) and mature (dried) maize kernels were sectioned in fixative (0.1 M cacodylate buffer/0.5% gluteraldehyde/2% paraformaldehyde). Tissue blocks were incubated in fixative for 2 h at 4°C and rinsed three times in 0.1 M cacodylate buffer, 15 min each wash, on a rotating shaker. This step was followed by a succession of dehydrating treatments as follows: tissue blocks were rinsed with 50% ethanol for 15 min, followed by incubation with 70% ethanol for 2 h at room temperature. The 70% ethanol was removed and replaced with 95% ethanol for a 2-h incubation, followed by three 2-h incubations with 100% ethanol. Tissue blocks were then incubated in a gradually increasing concentration of white London Resin (LR white), starting with 1:3, 2:1, 3:1 (vol:vol) LR white to ethanol for 8–12 h each time, and finally with 100% LR white overnight. Incubation in pure LR white was repeated twice for 8–12 h each time, after which the tissue blocks were cast in gelatin capsules for 24–48 h at 60°C to polymerize. Ultrathin sections were cut and mounted on grids for scanning transmission electron microscope by the Bessey Microscopy Facility at Iowa State University.
Immunogold Labeling. Kernel sections mounted on grids were incubated in TBS blocking buffer (0.05 M Tris, pH 8.3/0.85% NaCl, supplemented with 0.5% BSA/0.5% normal serum/3% dry nonfat milk) for 2 h at room temperature, followed by incubation of grids in primary antibodies, goat anti-LT-B polyclonal sera (Biogenesis, Bournewith, U.K.), rabbit anti-LT-B, rabbit anti-granule-bound starch synthase (GBSS) (S. Wessler, University of Georgia, Athens, GA), or rabbit anti-α-zein polyclonal sera at 37°C for 2 h. All primary antibodies were diluted 1:50 (vol:vol) in blocking buffer, with the exception of the rabbit antizein, which was diluted 1:100 (vol:vol), also in blocking buffer. Negative control grids were incubated in fresh blocking buffer. Grids were washed three times, 10 min each wash, in TBS buffer supplemented with 0.5% normal serum and 0.5% BSA, and incubated at room temperature in donkey anti-goat polyclonal serum or goat anti-rabbit polyclonal serum conjugated to 12 nm of gold, diluted 1:20 and 1:50 (vol:vol), respectively, in TBS buffer supplemented with 0.5% normal donkey (for goat anti-LT-B grids), or normal goat serum (for rabbit anti-GBSS, LT-B, or zein grids), and 0.5% BSA/0.1% fish gelatin. Finally, the grids were washed three times, 10 min each wash, in distilled water and allowed to air dry before viewing under the scanning transmission electron microscope.
Starch Sample Preparation. Starch was purified by using the procedure of White et al. (14), with the following modifications. Mature dry kernels were soaked in glass vials containing 2–4 ml of 0.45% Na2S2O5 for 3 d. The pericarp and embryo were removed by using a razor blade. Endosperms were placed into a 50-ml conical tube with 10 ml of distilled water and homogenized with a rotor stator homogenizer. Homogenate was vacuum filtered through a 30-μM nylon filter (Spectrum Laboratories, Houston, no. 146506) cut to size in a side-arm flask, and the filtrate collected in a 50-ml conical tube placed under the filter assembly. Filtered homogenates were allowed to settle for a minimum of 1.5 h at 4°C. Water was removed from the sample by aspiration and the remaining starch water slurry transferred to a 2-ml tube. The slurry was centrifuged at low speed for 5 min. The starch was again washed three times with 1.5 ml of water, and one of three reagents, 75% ethanol, 95% ethanol, or 75% ethanol plus 3% β-mercaptoethanol. Samples were dried in a centrifugal evaporator (Speed-Vac, Savant). To determine the amount of LT-B in each kernel fraction, mature P77 kernels from an individual ear were used. LT-B-expressing and -non-expressing segregants were distinguished and separated by endosperm drilling and subsequent GM1 ELISA, as described (3). Endosperm from both groups were split in three parts, one portion for starch extraction for determination of starch-associated LT-B, the other two for LT-B, and total protein determination in the whole endosperm fraction.
Protein Extraction and Measurements. Total proteins from kernel materials were extracted by using the following buffer: 25 mM sodium phosphate (pH 6.6)/100 mM NaCl/0.1% Triton X-100 (vol/vol)/1 mM EDTA/10 μg/ml of leupeptin (wt/vol)/0.25 mM serine protease inhibitor Perfabloc SC (Fluka), at room temperature. Aqueous extractable protein was determined in all samples (drilled kernels, crushed endosperms, and starch) by using the Bradford assay (15). Total nitrogen determination and protein content conversion were performed by using a LECO CHN-2000 (LECO, St. Joseph, MI), according to AOAC Official Method 990.03.
Removal of Exogenous Proteins. To remove exogenous proteins from purified starch or ground maize kernel, samples were incubated with 5 mM CaCl2 solution containing 100 μg/ml thermolysin (Sigma) at 64°C for ≈16 h (16). The reactions were terminated by the addition of EDTA to 20 mM. Samples were subsequently washed five times in distilled water to remove thermolysin and were then boiled in SDS sample buffer (17) (1 ml per 50 mg of starch) for 7 min at 95°C for Western analysis (3).
Determination of Thermal Stability. To determine the thermal stability of maize-expressed LT-B, 50 mg of ground meal from P77 transgenic maize meal was wetted with 250 μl of sterile distilled water, and an identical amount of nontransgenic maize meal was wetted in sterile water spiked with 1 μg/ml LT-B. The wet meals were incubated at room temperature (25°C), and 37, 55, 65, or 85°C for 2 or 4 h. At the end of the treatment, 250 μl of 2× protein extraction buffer was added to all samples, for a final concentration of 10 μl of extraction buffer for every milligram of ground maize meal. All samples were shaken for 1 h at room temperature, and functional LT-B was quantified by GM1 ELISA, as described. Resistance to thermal degradation was determined by comparing the functional LT-B remaining in the treated samples to the functional LT-B in the samples not subjected to any heat treatment.
Simulated Gastric Fluid (SGF) Digests. The SGF contained 3.2 mg/ml porcine pepsin (Sigma) in 34 mM NaCl, 0.7% HCl, as described (18). Twenty-five milligrams of transgenic maize meal containing 0.05 μg of functional LT-B was incubated in 250 μl of SGF. For controls, 5 μg of recombinant LT-B (J. Clements, Tulane University, New Orleans) was added to 25 mg of nontransgenic maize meal and also incubated in 250 μl of SGF. Incubations were done at 37°C for 30 s; 1, 5, 15, 30 min; or 2 h. The reactions were terminated by the addition of 50 μl of 1.5 M Tris·HCl (pH 8.8), 200 μl of 2× SDS sample buffer containing β-mercaptoethanol, and boiled at 95°C for 7 min. Fifty microliters of each treated sample was separated on an SDS/18% PAGE, transferred to a 0.45-μm nitrocellulose membrane, and analyzed by Western analysis (3).
Results
An sLT-B was fused to DNA sequences encoding either the native LT-B signal peptide (Construct P77) or maize 27 kDa γ-zein signal peptide (construct P81), as described in Fig. 1. Twenty independent transformation events from each construct were evaluated for the expression of the LT-B gene in seed by using a ganglioside-dependent enzyme-linked immunosorbent assay that measures functional LT-B (3). The LT-B proteins produced by maize plants carrying either construct (P77 and P81) were structurally and functionally identical to the bacterium-derived LT-B protein. Oral immunization with maize-synthesized LT-B induced a strong immune response in BALB/c mice, protecting them from challenge with the E. coli LT and its homologue cholera toxin (3).
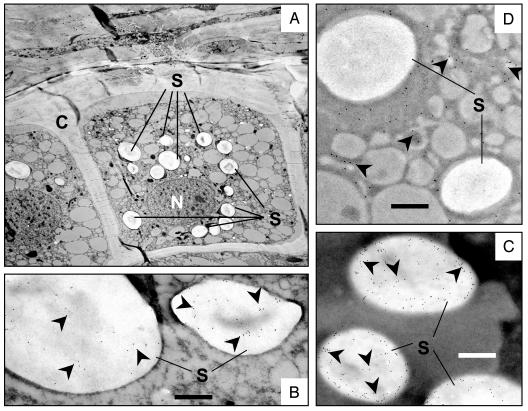
Immunogold labeling was used to determine the subcellular localization of LT-B within transgenic maize kernels. Ultrathin sections of 23-day-old immature kernels from P77 and nontransgenic control plants were processed as described in Materials and Methods and viewed by transmission electron microscopy. As shown in Fig. 2B, LT-B was detected exclusively in starch granules of immature kernels of P77 transgenic lines. No specific signal could be detected near or within cell walls, intercellular spaces, the endoplasmic reticulum (ER), Golgi apparatus, or secretory vesicles. No gold particles could be detected in the mature and immature kernels of nontransgenic maize and nontransgenic segregants (data not shown). For a positive control, we used the maize GBSS, an enzyme involved in starch biosynthesis and naturally found inside starch granules (19). Fig. 2C shows an identical localization pattern for the GBSS and LT-B, confirming the starch localization of LT-B in transgenic maize lines. To rule out the possibility of artifactual gold accumulation in the starch as a result of the labeling process, we used the same procedure to determine the localization of the maize α-zein protein as another control. The zein proteins are the major seed storage protein in maize, comprising 50–60% of endosperm protein (10), and are localized outside of the amyloplast, the subcellular compartment in which starch granules are formed. As shown in Fig. 2D, α-zein protein was localized exclusively in the cytoplasm of the cell, and no gold particles could be detected in the starch granules, indicating that gold localization in starch was not a result of the labeling process in our hands. The α-zein was expected to localize in intact protein bodies, and the cytoplasmic localization observed here shows disruption of the protein bodies during maturation and/or processing.
Fig. 2.
Immunolocalization of LT-B in immature maize kernels. (A) Section of maize kernel showing a cell beneath the aleurone layer. C, cell wall; N, nucleus; S, starch granules. (B) P77 transgenic kernels showing LT-B localization in starch granules. (C) GBSS localization in starch granules. (D) α-Zein protein localization in cytoplasm. Arrowheads indicate gold particles. (Bars = 500 nm at ×18,000.)
To investigate whether the starch localization of LT-B protein requires its native signal peptide, immunolocalization of LT-B was carried out in LT-B-expressing P81 transgenic maize kernels, in which the bacterial signal peptide was replaced with the signal peptide from maize γ-zein protein (Fig. 1). As observed in P77 kernels, starch granule localization of LT-B was also detected in P81 kernels (data not shown). This result indicated that the native signal peptide of LT-B was not necessary for the starch localization of the LT-B protein in transgenic maize seed.
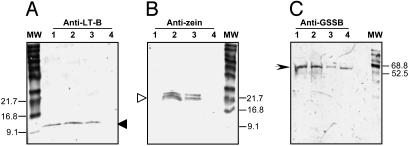
To determine whether the LT-B proteins were localized at the surface or inside of the starch granules, starch samples from mature transgenic seeds were collected and treated to remove surface proteins, and granule-associated proteins were analyzed on immunoblots. The approach was based on the fact that polypeptides within the starch granules are not susceptible to hydrolysis on treatment of intact granules with exogenous proteases (16). Starch samples were isolated from individual mature kernels of the transgenic maize line carrying construct P77, using three different levels of preparation stringency (75% ethanol, 95% ethanol, or 75% ethanol plus 3% β-mercaptoethanol) to remove external proteins. The starch samples were then treated with thermolysin (EC 3.4.24.27) for 16 h to remove remaining external proteins not embedded in the starch granules. The samples were then boiled to disrupt the starch granules, releasing embedded proteins. Duplicate SDS/PAGE gels containing equal amounts of protein were analyzed by Western blotting by using antibodies against LT-B, GBSS, or α-zein proteins. Zeins serve as convenient markers for extragranular protein contamination, whereas GBSS is an internal control for starch-bound proteins. As shown in Fig. 3, both LT-B and zein proteins could be detected in starch samples treated with 75 or 95% ethanol (lanes 2 and 3 of Fig. 3 A and B). However, only LT-B protein could be detected in starch samples subjected to the most stringent treatment with 75% ethanol/3% β-mercaptoethanol (lane 1, Fig. 3A). Neither protein was detected in the nontransgenic starch samples treated the same way (lane 4 in Fig. 3 A and B). Western analysis also shows the presence of the 60-kDa GBSS used as the positive control for starch-encapsulated proteins in all boiled and protease-treated samples, as expected (Fig. 3C, lanes 1–4). These results support the immunogold localization observation that LT-B is present within the starch granules of maize kernels.
Fig. 3.
Western blot analyses of total proteins from starch samples after thermolysin treatment. The samples were separated on a SDS/12% PAGE, transferred to a 0.45-μm nitrocellulose membrane, and probed with goat anti-LT-B antibodies (A), rabbit anti-zein antibodies (B), or rabbit anti-GBSS (C), followed by rabbit anti-goat or goat anti-rabbit alkaline phosphatase conjugate, respectively. P77 starch samples washed with 75% ethanol plus 3% β-mercaptoethanol (lane 1), 75% ethanol (lane 2), or 95% ethanol (lane 3), and B73 nontransgenic starch samples washed with 75% ethanol plus 3% β-mercaptoethanol (lane 4), respectively. Solid arrows in A, monomeric form of LT-B (11.6 kDa); open arrow in B, zein proteins (19 and 22 kD); arrowhead in C, GBSS (60 kD).
Although LT-B protein was consistently detected in starch granules by two different sets of antibodies (goat anti-LT-B and rabbit anti-LT-B followed by the relevant secondary antibody gold conjugates), Western blots and GM1-dependent ELISA analysis of ground kernels (3) and intact starch granules indicated that the LT-B protein is also present in the soluble fraction of the endosperm. These observations suggested that LT-B has similar properties to some starch biosynthetic enzymes, such as starch synthase I, which are present in both soluble and granule-bound forms in amyloplast of maize endosperm (19). We conducted further analyses of the distribution of LT-B between the starch fraction and the soluble fraction of the endosperm.
Table 1 shows the partitioning of total protein and LT-B between starch and the soluble fraction. Total protein determined by the combustion analysis method was 8.784% of ground endosperm material, in the range of average protein content (8.7%) in endosperm of a normal dent maize variety (20). Total aqueous extractable protein (TAEP) determined by Bradford assay was 0.23% of the ground endosperm. The functional pentameric form of LT-B determined by the GM1-dependent ELISA is 0.0002% of the ground endosperm. The total protein determined in the starch fraction was 2.028% of the dry starch weight, ≈4-fold reduction from the 8.784% observed in the ground endosperm. It is also noted that the ratio of total protein vs. TAEP in starch (2.028 vs. 0.027) is 75-fold, whereas this ratio in endosperm (8.784 vs. 0.234) is 37-fold. However, in the starch fraction, despite the significantly smaller amount of TAEP, LT-B constitutes 0.00013% of the dry weight, more than half the amount observed in the whole endosperm fraction (0.0002%). These data are a quantitative indication that more than half of the LT-B in the endosperm is starch associated.
Table 1. Percentage of total protein and LT-B in transgenic maize kernel fractions.
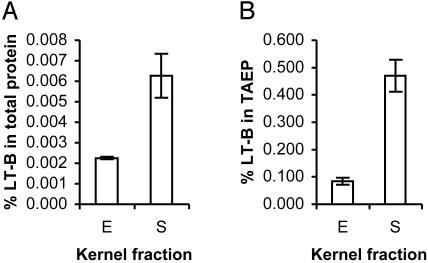
Fig. 4 illustrates that LT-B is detectable in the soluble fraction of endosperm cells prepared without disrupting starch granule structure, and that it is also externally associated with the purified starch sample. Fig. 4A shows that LT-B constitutes a much larger proportion of total protein in starch (>0.006%) than in endosperm (>0.002%). A similar observation was made on TAEP. Fig. 4B shows that LT-B constitutes >0.45% of TAEP in starch, whereas in endosperm, LT-B is <0.1% of TAEP. These observations are indicative of a strong association between LT-B and starch in endosperm tissue.
Fig. 4.
Percentage of LT-B in total protein (A) and TAEP (B) of maize kernel fractions. E, endosperm fraction; S, starch fraction.
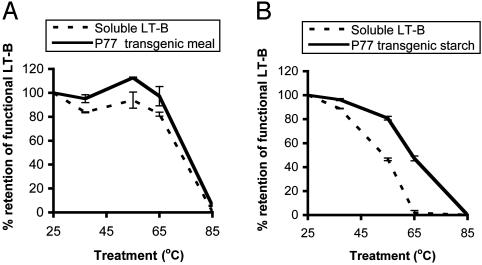
To determine the practical importance of the intricate association of LT-B with the maize tissue matrix, we examined its thermostability and resistance to proteolytic degradation by the biologically relevant gastric enzymes and small intestinal enzymes. It has been demonstrated that the B subunit dissociates between 66 and 78°C (5). Gelatinization of starch is an essential procedure in maize processing for humans and occurs above 65°C, depending on the length of exposure. We incubated wet transgenic maize meal and starch at 37, 55, 65, or 85°C for 2 or 4 h. Soluble bacterium-derived LT-B was added to an equal amount of nontransgenic maize meal or starch and subjected to the same treatment. Percent retention of functional LT-B in treated samples was established from the ratio of GM1-captured LT-B in treated to untreated samples (room temperature, 25°C). Fig. 5 shows the results after incubation of samples for 4 h; the data for 2-h incubation were similar but are not presented. Fig. 5A shows that functional LT-B increases with increase in incubation temperature up to 55°C. This upward trend could possibly be a result of increased release of LT-B from the corn matrix. These data are consistent with previous observations that incubation of transgenic maize meal at elevated temperature enhanced LT-B extraction (R.K.C., unpublished work). Functional LT-B declines at 65°C; LT-B in ground meal declines slightly to 97.2%, whereas the soluble LT-B declines more markedly to 82.1% (Fig. 5A). At 85°C, 5.49% functional LT-B is retained in the transgenic meal, but no functional LT-B can be detected in the soluble LT-B samples. Similar trends in functional LT-B retention were observed in starch (Fig. 5B). Transgenic starch appears to have a similar protective effect on LT-B to the transgenic maize meal. This effect is markedly pronounced when the starch samples were incubated at gelatinization temperatures (65 or 85°C) for both 2-(data not shown) and 4-h treatments, with a much higher protective effect apparent after incubation for the 4-h period (Fig. 5B). In starch samples, however, no functional LT-B is detected after incubation at 85°C for 4 h. It is noted that at temperatures above 65°C, the purified starch gelatinizes more readily than the whole meal. The retention of functional LT-B in its functional form is therefore achieved in both transgenic starch and transgenic whole meal but is stronger in the latter.
Fig. 5.
Retention of functional (GM1-binding) LT-B in TAEP after 4 h of incubation at 37, 55, 65, and 85°C. (A) Ground maize meal. (B) Purified maize starch. Solid lines, transgenic maize line P77; dotted lines, nontransgenic maize control spiked with 1 μg/ml of bacterium-derived soluble LT-B.
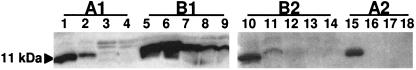
Fig. 6 shows the results of Western blot analysis indicating the presence of monomeric LT-B in samples after SGF treatment. Twenty-five milligrams of transgenic maize material contained 0.05 μg of functional pentameric LT-B determined by GM1-dependent ELISA (Table 1, Fig. 6 B1 and B2). The control was 25 mg of nontransgenic maize material, to which 5 μg of bacterial LT-B was added (Fig. 6 A1 and A2). Soluble LT-B added to nontransgenic maize meal was completely digested by pepsin within 5 min of SGF treatment (Fig. 6, lanes 2, 3, 4, and 16), whereas LT-B present in transgenic maize meal resisted SGF digestion up to 15 min (Fig. 6, lanes 6–9 and 11). It is important to note that we were not able to determine the exact amount of monomeric LT-B in transgenic maize material and bacterial LT-B control. However, it was reported that pentameric form of LT-B constituted ≈40% of total LT-B protein (2). Thus, we deduced there were ≈2 μg of pentameric LT-B in the control samples, 40× more LT-B compared with transgenic material.
Fig. 6.
Western blot analysis of LT-B after gastric fluid digestion simulation. (A1 and A2) Ground maize kernels mixed with bacterial LT-B. (B1 and B2) Ground transgenic maize kernels. The samples were treated with pepsin at various length of period and separated on an SDS/18% PAGE and transferred to a 0.45-μm nitrocellulose membrane and probed with rabbit anti-LT-B antibodies. Lanes 1, 5, 10, and 15, 0-min digestion (no digestion control); lanes 2 and 6, 30-s digestion; lanes 3 and 7, 1-min digestion; lanes 4 and 8, 5-min digestion; lanes 9, 11, and 16, 15-min digestion; lanes 12 and 17, 30-min digestion; lanes 13 and 18, 60-min digestion; lane 14, 120-min digestion.
The result of trypsin and chymotrypsin digests also showed that LT-B in general was resistant to degradation by small intestinal enzymes, but maize-associated LT-B was more resistant to these enzymes (data not shown). These data, together with our immunological results obtained from animal studies comparing maize-derived LT-B with bacterial LT-B (3), strongly suggest that the maize starch-associated LT-B has significant resistance to gastric enzymatic digestions and led to increased antibodies responses in mice (3).
Discussion
We observed in this work the localization of a bacterial protein in the starch granules of transgenic maize seed. These observations are contrary to our expectation, in which LT-B protein in transgenic maize kernels would be strictly ER targeted and ultimately secreted to the cell wall and extracellular spaces (21). This result led to further analyses of LT-B partitioning in maize endosperm. Data from these analyses strongly suggested that LT-B protein was detected not only internally but also externally of starch granules in maize endosperm. Interestingly, the immunogold localization did not reveal LT-B localized outside starch granules. This discrepancy between the immunogold-labeling observation and the soluble extractable protein ELISA as well as Western analysis data could be explained by the possibility that antibody-reactive epitopes of LT-B in the soluble kernel fraction were sensitive to the processing procedures and/or reagents, whereas the granule-bound forms were protected from fixative and polymerization procedures of tissue preparation. Another possibility is that the LT-B protein level in the soluble fraction of the endosperm was beneath the sensitivity level of the immunogold detection technique.
Our immunolocalization observations suggest that a novel amyloplast-targeting mechanism may be present in plant cells. In enterotoxigenic forms of E. coli, LT-B is targeted to the periplasmic space (22), whereas in yeast, LT-B was shown to be ER targeted, assembled into pentamers, and retained within the endomembrane system (23). In plants, the default pathway for proteins transported through the ER is secretion, although a protein's ultimate subcellular localization also depends on factors other than just the presence of a signal sequence, including topological information on the protein itself (24).
Although the import of nucleus-encoded proteins into chloroplasts has been studied extensively (25), the mechanism of import of nucleus-encoded proteins into amyloplasts has not been established (16). Chloroplast-targeted proteins are synthesized with transit peptides usually removed upon import into the chloroplast. Targeting to amyloplasts is thought to occur by a similar mechanism, because most nucleus-encoded starch biosynthetic enzymes have sequences reminiscent of chloroplast transit peptides (26).
The mechanism by which LT-B is sorted to starch granules is not clear. Judging by the migration of LT-B in SDS/PAGE, we believe maize-derived mature LT-B protein produced with either the bacterial signal peptide or the plant signal peptide was processed appropriately in plant cells (3) (data not shown). Because the maize-derived LT-B with either signal peptide was detected in starch granules, it seems likely that the starch targeting information must be contained on the mature LT-B protein itself. Current models of plastid targeting involve translation in the cytoplasm and subsequent import into the plastid. If LT-B is transported to starch granules via the ER, a mechanism for returning LT-B to the cytoplasm or a novel vesicle-mediated sorting pathway would be required. Transport of LT-B from the ER to the cytoplasm could involve retrotranslocation, whereby proteins are exported from the lumen or membrane of the ER into the cytosol (27). Retrotranslocation has been reported for the A subunits of the cholera and E. coli toxins (27–29). Whether the B subunits undergo a similar process is not determined.
Recently, the twin arginine translocase (tat) has been described, and this pathway commonly operates in eukaryotic thylakoid membranes and the prokaryotic plasma membranes (30). The novelty of this pathway is its unique ability to transport folded proteins through tightly sealed membranes without the hydrolysis of nucleotide triphosphate(s) at any stage in the translocation process. Proteins transported via this pathway have an essential twin-arginine motif in their signal peptides. The LT-B protein with or without the signal peptide motif does not have this feature. However, not all of the components of this pathway have been identified. The tat pathway has been shown to translocate multimeric proteins across membranes. It is possible that LT-B, a pentameric protein, is transported into the amyloplast by using a similar mechanism.
Few transgenic proteins have been analyzed in great detail for their subcellular localization in plants. Düring et al. (31) targeted immunoglobulin chains to intercellular spaces in transgenic tobacco by using the barley aleurone α-amylase signal peptide. This group observed localization of the assembled monoclonal antibodies in the ER and, unexpectedly, in the chloroplasts. Because no particular features of the immunoglobulins were expected to target the chloroplasts, they could not explain this observation. Similarly, our group observed LT-B localization in starch granules. It is possible that this association is facilitated by the affinity of LT-B to the galactose portion of the galactolipids on plastid membranes, because LT-B has a demonstrated affinity for galactose (5). The apparent starch localization of LT-B could well be random or explained by any of the pathways we describe. However, such unexpected observations hint at pathways and biochemical processes yet to be unraveled. Further investigation of the starch-targeting property of the LT-B protein by using site-directed mutagenesis and reporter genes will contribute to the elucidation of mechanisms of protein translocation to the plastid.
There are several apparent practical advantages of expressing LT-B in maize endosperm. First, our data (Table 1) indicate that in this transgenic line, 1.3 and 2 g of LT-B can be obtained from 1 kg of starch and endosperm, respectively. Because most aqueous soluble proteins are lost in the starch purification process, the large amount of soluble LT-B copurified with starch fraction suggested a tight association of LT-B with starch. The copurification of LT-B with starch can reduce the presence of other undesirable endosperm proteins in the final product. Second, the internal and external localization of LT-B protein with maize starch granules has important implications on the effectiveness of a maize-based oral vaccine. Our work demonstrates that expression of LT-B in transgenic maize endosperm tissue conferred heat resistance at temperature and exposure periods where the starch is partially or completely gelatinized, and gelatinization of starch is part of the processing treatments to make maize meal more palatable for humans, such as in the manufacture of breakfast cereals and other snacks. In this work, the purified starch gelatinizes more readily than the whole meal, hence the increased reduction in functional LT-B retention in transgenic kernel-derived starch compared with the ground transgenic meal. Although it is not clear whether the starch-internalized LT-B is in multimeric (functional) or monomeric form, we showed earlier that mice orally immunized with this LT-B-expressing transgenic maize meal developed a higher serum and mucosal antibody response than mice immunized with nontransgenic maize meal mixed with an equivalent amount of bacterial LT-B (3). One of the major hurdles to oral immunization is the fact that antigens have to survive the severely low pH and degradative action of the gastric environment (32). We demonstrated here that maize-derived LT-B is more resistant to peptic cleavage in SGF digestion analysis. Vaccines encapsulated in or strongly bound to starch granules could be protected from the harsh environment of the stomach and possibly released slowly from the starch, resulting in prolonged exposure of the antigen to the gut-associated mucosal system. In addition, encapsulation of novel proteins in starch by using a domain in the LT-B protein as a targeting signal could greatly facilitate the protein extraction and purification process in maize.
Acknowledgments
We thank Tracy Pepper and Jack Horner for electron microscopy services, John Clements for technical advice and supply of LT-B, Erik Mottl for assistance with starch preparation, Sue Wessler for providing anti-GBSS antibody, Jennifer McMurray for technical assistance, and Martha James for discussion and critical reviewing of the manuscript. Transgenic maize plants were generated in the Plant Transformation Facility, and total protein content measurement was performed in the Soil and Plant Analysis Laboratory of Department of Agronomy at Iowa State University. We thank the Rockefeller Foundation for providing a graduate study fellowship (to R.K.C.). This work was supported by United States Department of Agriculture–National Research Initiative Grant 99-35504-7799.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: LT-B, Escherichia coli heat labile enterotoxin B subunit; sLT-B, synthetic LT-B gene; SGF, simulated gastric fluid; ER, endoplasmic reticulum; GBSS, granule-bound starch synthase; TAEP, total aqueous extractable protein.
References
- 1.Mason, H. S., Haq, T. A., Clements, J. D. & Arntzen, C. J. (1998) Vaccine 16, 1336-1343. [DOI] [PubMed] [Google Scholar]
- 2.Streatfield, S. J., Jilka, J. M., Hood, E. E., Turner, D. D., Bailey, M. R., Mayor, J. M., Woodard, S. L., Beifuss, K. K., Horn, M. E., Delaney, D. E., et al. (2001) Vaccine 19, 2742-2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chikwamba, R., Cunnick, J., Hathaway, D., McMurray, J., Mason, H. & Wang, K. (2002) Transgenic Res. 11, 479-493. [DOI] [PubMed] [Google Scholar]
- 4.Millar, D. G., Hirst, T. R. & Snider, D. P. (2001) Infect. Immun. 69, 3476-3482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Spangler, B. D. (1992) Microbiol. Rev. 56, 622-647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fujiwara, Y. & Asogawa, M. (2001) Genome Inform. Ser. Workshop Genome Inform. 12, 103-112. [PubMed] [Google Scholar]
- 7.Chapman, R., Sidrauski, C. & Walter, P. (1998) Annu. Rev. Cell Dev. Biol. 14, 459-485. [DOI] [PubMed] [Google Scholar]
- 8.Van den Broeck, G., Timko, M. P., Kausch, A. P., Cashmore, A. R., Van Montagu, M. & Herrera-Estrella, L. (1985) Nature 313, 358-363. [DOI] [PubMed] [Google Scholar]
- 9.Bruce, B. D. (2000) Trends Cell Biol. 10, 440-447. [DOI] [PubMed] [Google Scholar]
- 10.Marks, M. D., Lindell, J. S. & Larkins, B. A. (1985) J. Biol. Chem. 260, 16445-16450. [PubMed] [Google Scholar]
- 11.Gallie, D. R., Tanguay, R. L. & Leathers, V. (1995) Gene 165, 233-238. [DOI] [PubMed] [Google Scholar]
- 12.Mason, H. S., DeWald, D. B. & Mullet, J. E. (1993) Plant Cell 5, 241-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frame, B. R., Zhang, H., Cocciolone, S. M., Sidorenko, L. V., Dietrich, C. R., Pegg, S. E., Zhen, S., Schnable, P. S. & Wang, K. (2000) In Vitro Cell. Dev. Biol. Plant 36, 21-29. [Google Scholar]
- 14.White, P., I. Abbas, L. Pollak, & Johnson, L. (1990) Cereal Chem. 67, 70-73. [Google Scholar]
- 15.Bradford, M. M. (1976) Anal. Biochem. 72, 248-254. [DOI] [PubMed] [Google Scholar]
- 16.Mu-Forster, C. & Wasserman, B. P. (1998) Plant Physiol. 116, 1563-1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Laemmli, U. K. (1970) Nature 227, 680-685. [DOI] [PubMed] [Google Scholar]
- 18.Roesler, K. R. & Rao, A. G. (2001) J. Agric. Food Chem. 49, 3443-3451. [DOI] [PubMed] [Google Scholar]
- 19.Mu-Forster, C., Huang, R., Powers, J. R., Harriman, R. W., Knight, M., Singletary, G. W., Keeling, P. L. & Wasserman, B. P. (1996) Plant Physiol. 111, 821-829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnson, L. A. (2000) in Handbook of Cereal Science and Technology, eds. Kulp, K. & Ponte, J. G., Jr. (Dekker, New York), pp. 31-80.
- 21.Ellgaard, L., Molinari, M. & Helenius, A. (1999) Science 286, 1882-1888. [DOI] [PubMed] [Google Scholar]
- 22.Hirst, T. R., Hardy, S. J. & Randall, L. L. (1983) J. Bacteriol. 153, 21-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schonberger, O., Hirst, T. R. & Pines, O. (1991) Mol. Microbiol. 5, 2663-2671. [DOI] [PubMed] [Google Scholar]
- 24.Stachelin, L. A. & Moore, I. (1995) Annu. Rev. Plant Physiol. Plant Mol. Biol. 46, 261-288. [Google Scholar]
- 25.Keegstra, K. & Cline, K. (1999) Plant Cell 11, 557-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mason-Gamer, R. J., Weil, C. F. & Kellogg, E. A. (1998) Mol. Biol. Evol. 15, 1658-1673. [DOI] [PubMed] [Google Scholar]
- 27.Tsai, B., Ye, Y. & Rapoport, T. A. (2002) Nat. Rev. Mol. Cell. Biol. 3, 246-255. [DOI] [PubMed] [Google Scholar]
- 28.Lord, J. M. & Roberts, L. M. (1998) J. Cell Biol. 140, 733-736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sandvig, K. & van Deurs, B. (2000) EMBO J. 19, 5943-5950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Robinson, C. & Bolhuis, A. (2001) Nat. Rev. Mol. Cell. Biol. 2, 350-356. [DOI] [PubMed] [Google Scholar]
- 31.Düring, K., Hippe, S., Kreuzaler, F. & Schell, J. (1990) Plant Mol. Biol. 15, 281-293. [DOI] [PubMed] [Google Scholar]
- 32.Cripps, A. W., Kyd, J. M. & Foxwell, A. R. (2001) Vaccine 19, 2513-2515. [DOI] [PubMed] [Google Scholar]