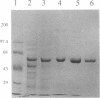
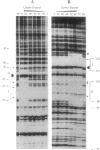
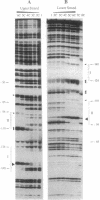
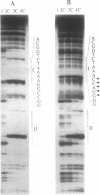
Abstract
The enteric NtrC (NRI) protein has been the paradigm for a class of bacterial enhancer-binding proteins (EBPs) that activate transcription of RNA polymerase containing the sigma 54 factor. Activators in the NtrC class are characterized by essentially three properties: (i) they bind to sites distant from the promoters that they activate (> 100 bp upstream of the transcriptional start site), (ii) they contain a conserved nucleotide-binding fold and exhibit ATPase activity that is required for activation, and (iii) they activate the sigma 54 RNA polymerase. We have characterized the NtrC protein from a photosynthetic bacterium, Rhodobacter capsulatus, which represents a metabolically versatile group of bacteria found in aquatic environments. We have shown that the R. capsulatus NtrC protein (RcNtrC) binds to two tandem sites that are distant from promoters that it activates, nifA1 and nifA2. These tandem binding sites are shown to be important for RcNtrC-dependent nitrogen regulation in vivo. Moreover, the conserved nucleotide-binding fold of RcNtrC is required to activate nifA1 and nifA2 but is not required for DNA binding of RcNtrC to upstream activation sequences. However, nifA1 and nifA2 genes do not require the sigma 54 for activation and do not contain the highly conserved nucleotides that are present in all sigma 54-type, EBP-activated promoters. Thus, the NtrC from this photosynthetic bacterium represents a novel member of the class of bacterial EBPs. It is probable that this class of EBPs is more versatile in prokaryotes than previously envisioned.
Full text
PDF









Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Austin S., Dixon R. The prokaryotic enhancer binding protein NTRC has an ATPase activity which is phosphorylation and DNA dependent. EMBO J. 1992 Jun;11(6):2219–2228. doi: 10.1002/j.1460-2075.1992.tb05281.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin S., Kundrot C., Dixon R. Influence of a mutation in the putative nucleotide binding site of the nitrogen regulatory protein NTRC on its positive control function. Nucleic Acids Res. 1991 May 11;19(9):2281–2287. doi: 10.1093/nar/19.9.2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avtges P., Kranz R. G., Haselkorn R. Isolation and organization of genes for nitrogen fixation in Rhodopseudomonas capsulata. Mol Gen Genet. 1985;201(3):363–369. doi: 10.1007/BF00331324. [DOI] [PubMed] [Google Scholar]
- Bender R. A. The role of the NAC protein in the nitrogen regulation of Klebsiella aerogenes. Mol Microbiol. 1991 Nov;5(11):2575–2580. doi: 10.1111/j.1365-2958.1991.tb01965.x. [DOI] [PubMed] [Google Scholar]
- Beynon J., Cannon M., Buchanan-Wollaston V., Cannon F. The nif promoters of Klebsiella pneumoniae have a characteristic primary structure. Cell. 1983 Sep;34(2):665–671. doi: 10.1016/0092-8674(83)90399-9. [DOI] [PubMed] [Google Scholar]
- Birkmann A., Böck A. Characterization of a cis regulatory DNA element necessary for formate induction of the formate dehydrogenase gene (fdhF) of Escherichia coli. Mol Microbiol. 1989 Feb;3(2):187–195. doi: 10.1111/j.1365-2958.1989.tb01807.x. [DOI] [PubMed] [Google Scholar]
- Buck M., Cannon W. Specific binding of the transcription factor sigma-54 to promoter DNA. Nature. 1992 Jul 30;358(6385):422–424. doi: 10.1038/358422a0. [DOI] [PubMed] [Google Scholar]
- Buikema W. J., Szeto W. W., Lemley P. V., Orme-Johnson W. H., Ausubel F. M. Nitrogen fixation specific regulatory genes of Klebsiella pneumoniae and Rhizobium meliloti share homology with the general nitrogen regulatory gene ntrC of K. pneumoniae. Nucleic Acids Res. 1985 Jun 25;13(12):4539–4555. doi: 10.1093/nar/13.12.4539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon W., Buck M. Central domain of the positive control protein NifA and its role in transcriptional activation. J Mol Biol. 1992 May 20;225(2):271–286. doi: 10.1016/0022-2836(92)90921-6. [DOI] [PubMed] [Google Scholar]
- Claverie-Martin F., Magasanik B. Role of integration host factor in the regulation of the glnHp2 promoter of Escherichia coli. Proc Natl Acad Sci U S A. 1991 Mar 1;88(5):1631–1635. doi: 10.1073/pnas.88.5.1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collado-Vides J., Magasanik B., Gralla J. D. Control site location and transcriptional regulation in Escherichia coli. Microbiol Rev. 1991 Sep;55(3):371–394. doi: 10.1128/mr.55.3.371-394.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contreras A., Drummond M. The effect on the function of the transcriptional activator NtrC from Klebsiella pneumoniae of mutations in the DNA-recognition helix. Nucleic Acids Res. 1988 May 11;16(9):4025–4039. doi: 10.1093/nar/16.9.4025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon R. Tandem promoters determine regulation of the Klebsiella pneumoniae glutamine synthetase (glnA) gene. Nucleic Acids Res. 1984 Oct 25;12(20):7811–7830. doi: 10.1093/nar/12.20.7811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond M. H., Contreras A., Mitchenall L. A. The function of isolated domains and chimaeric proteins constructed from the transcriptional activators NifA and NtrC of Klebsiella pneumoniae. Mol Microbiol. 1990 Jan;4(1):29–37. doi: 10.1111/j.1365-2958.1990.tb02012.x. [DOI] [PubMed] [Google Scholar]
- Ferro-Luzzi Ames G., Nikaido K. Nitrogen regulation in Salmonella typhimurium. Identification of an ntrC protein-binding site and definition of a consensus binding sequence. EMBO J. 1985 Feb;4(2):539–547. doi: 10.1002/j.1460-2075.1985.tb03662.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster-Hartnett D., Cullen P. J., Gabbert K. K., Kranz R. G. Sequence, genetic, and lacZ fusion analyses of a nifR3-ntrB-ntrC operon in Rhodobacter capsulatus. Mol Microbiol. 1993 May;8(5):903–914. doi: 10.1111/j.1365-2958.1993.tb01636.x. [DOI] [PubMed] [Google Scholar]
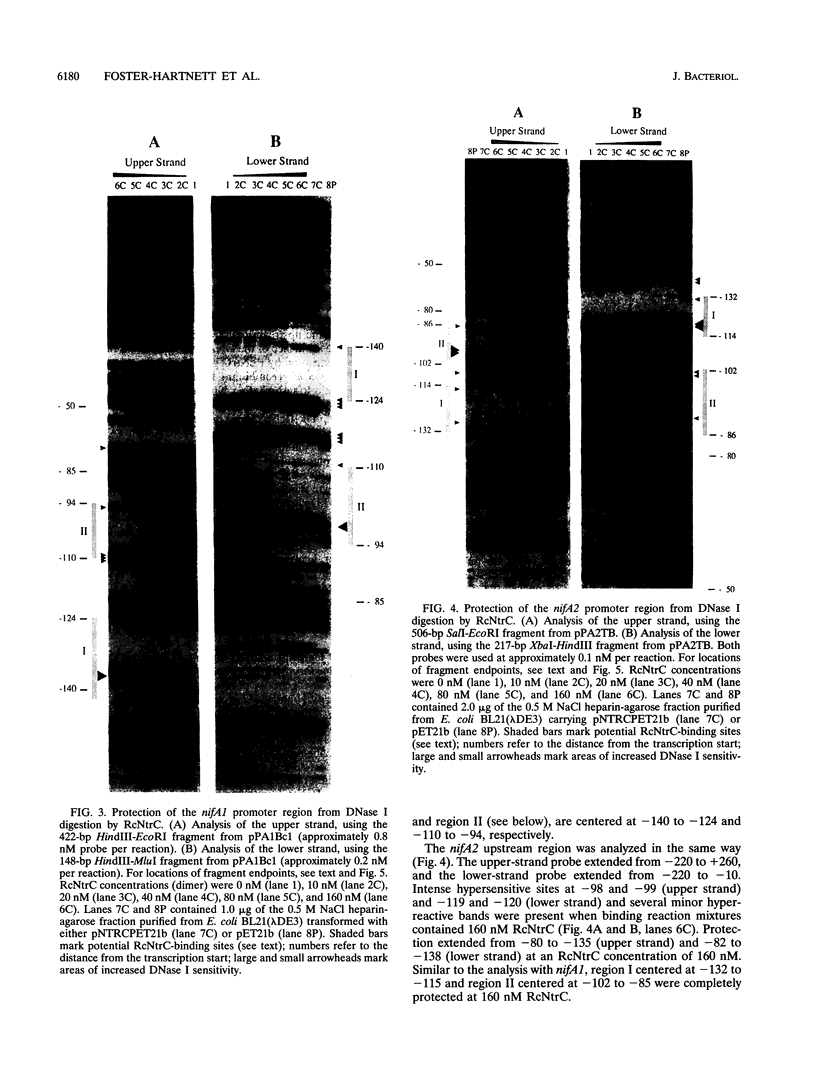
- Foster-Hartnett D., Kranz R. G. Analysis of the promoters and upstream sequences of nifA1 and nifA2 in Rhodobacter capsulatus; activation requires ntrC but not rpoN. Mol Microbiol. 1992 Apr;6(8):1049–1060. doi: 10.1111/j.1365-2958.1992.tb02170.x. [DOI] [PubMed] [Google Scholar]
- Foster-Hartnett D., Kranz R. G. The Rhodobacter capsulatus glnB gene is regulated by NtrC at tandem rpoN-independent promoters. J Bacteriol. 1994 Aug;176(16):5171–5176. doi: 10.1128/jb.176.16.5171-5176.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galas D. J., Schmitz A. DNAse footprinting: a simple method for the detection of protein-DNA binding specificity. Nucleic Acids Res. 1978 Sep;5(9):3157–3170. doi: 10.1093/nar/5.9.3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gralla J. D. Transcriptional control--lessons from an E. coli promoter data base. Cell. 1991 Aug 9;66(3):415–418. doi: 10.1016/0092-8674(81)90001-5. [DOI] [PubMed] [Google Scholar]
- Gussin G. N., Ronson C. W., Ausubel F. M. Regulation of nitrogen fixation genes. Annu Rev Genet. 1986;20:567–591. doi: 10.1146/annurev.ge.20.120186.003031. [DOI] [PubMed] [Google Scholar]
- Hawley D. K., McClure W. R. Compilation and analysis of Escherichia coli promoter DNA sequences. Nucleic Acids Res. 1983 Apr 25;11(8):2237–2255. doi: 10.1093/nar/11.8.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschman J., Wong P. K., Sei K., Keener J., Kustu S. Products of nitrogen regulatory genes ntrA and ntrC of enteric bacteria activate glnA transcription in vitro: evidence that the ntrA product is a sigma factor. Proc Natl Acad Sci U S A. 1985 Nov;82(22):7525–7529. doi: 10.1073/pnas.82.22.7525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoover T. R., Santero E., Porter S., Kustu S. The integration host factor stimulates interaction of RNA polymerase with NIFA, the transcriptional activator for nitrogen fixation operons. Cell. 1990 Oct 5;63(1):11–22. doi: 10.1016/0092-8674(90)90284-l. [DOI] [PubMed] [Google Scholar]
- Hübner P., Willison J. C., Vignais P. M., Bickle T. A. Expression of regulatory nif genes in Rhodobacter capsulatus. J Bacteriol. 1991 May;173(9):2993–2999. doi: 10.1128/jb.173.9.2993-2999.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inouye S., Nakazawa A., Nakazawa T. Nucleotide sequence of the regulatory gene xylR of the TOL plasmid from Pseudomonas putida. Gene. 1988 Jun 30;66(2):301–306. doi: 10.1016/0378-1119(88)90366-6. [DOI] [PubMed] [Google Scholar]
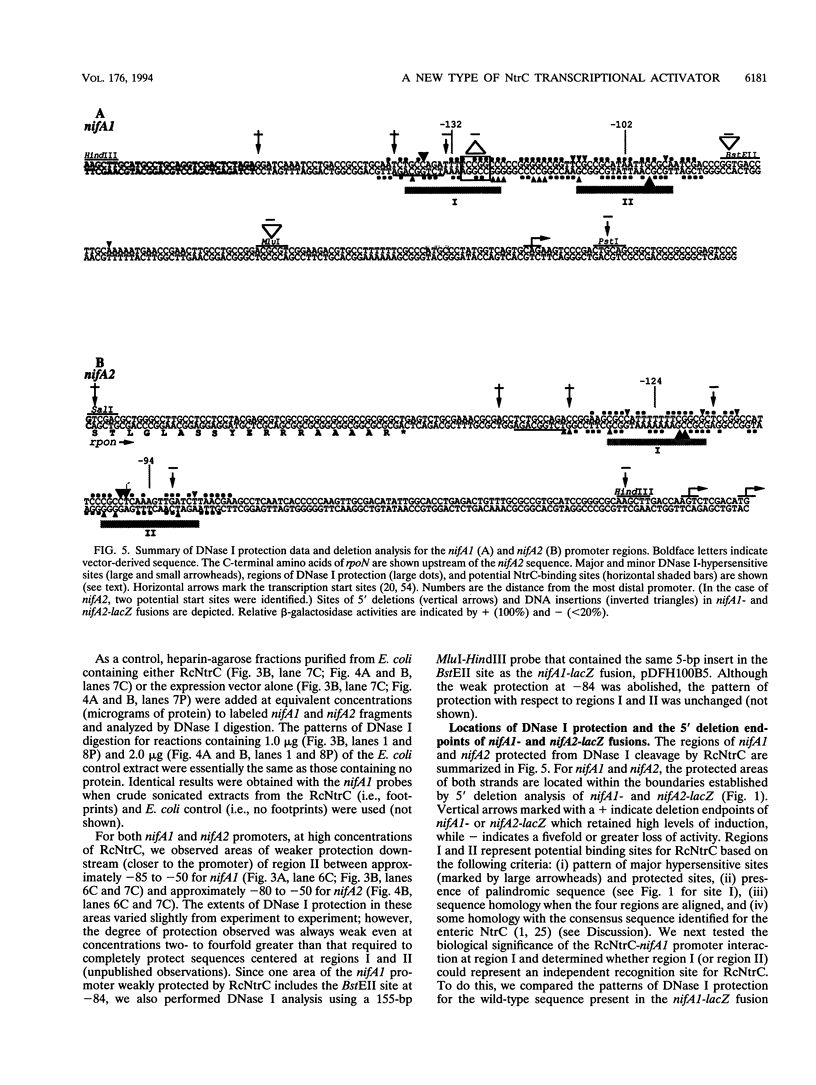
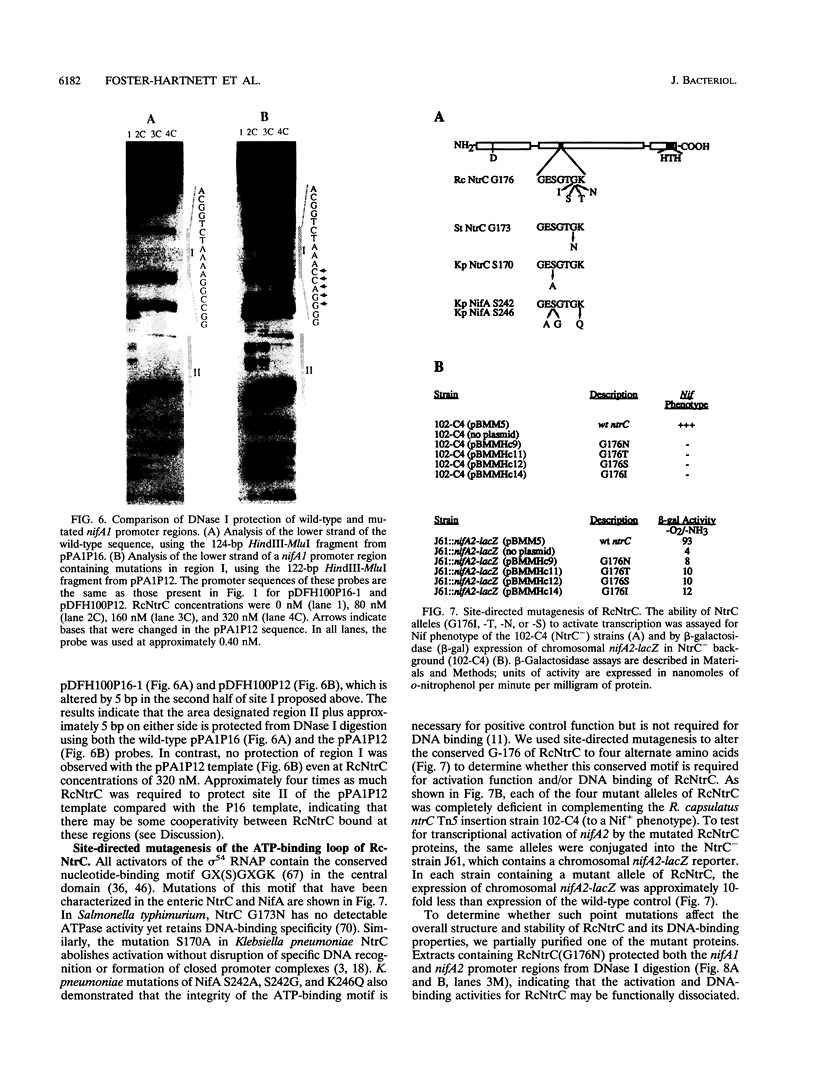
- Jones R., Haselkorn R. The DNA sequence of the Rhodobacter capsulatus ntrA, ntrB and ntrC gene analogues required for nitrogen fixation. Mol Gen Genet. 1989 Feb;215(3):507–516. doi: 10.1007/BF00427050. [DOI] [PubMed] [Google Scholar]
- Keener J., Kustu S. Protein kinase and phosphoprotein phosphatase activities of nitrogen regulatory proteins NTRB and NTRC of enteric bacteria: roles of the conserved amino-terminal domain of NTRC. Proc Natl Acad Sci U S A. 1988 Jul;85(14):4976–4980. doi: 10.1073/pnas.85.14.4976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kranz R. G., Haselkorn R. Characterization of nif regulatory genes in Rhodopseudomonas capsulata using lac gene fusions. Gene. 1985;40(2-3):203–215. doi: 10.1016/0378-1119(85)90043-5. [DOI] [PubMed] [Google Scholar]
- Kranz R. G., Pace V. M., Caldicott I. M. Inactivation, sequence, and lacZ fusion analysis of a regulatory locus required for repression of nitrogen fixation genes in Rhodobacter capsulatus. J Bacteriol. 1990 Jan;172(1):53–62. doi: 10.1128/jb.172.1.53-62.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kustu S., North A. K., Weiss D. S. Prokaryotic transcriptional enhancers and enhancer-binding proteins. Trends Biochem Sci. 1991 Nov;16(11):397–402. doi: 10.1016/0968-0004(91)90163-p. [DOI] [PubMed] [Google Scholar]
- Kustu S., Santero E., Keener J., Popham D., Weiss D. Expression of sigma 54 (ntrA)-dependent genes is probably united by a common mechanism. Microbiol Rev. 1989 Sep;53(3):367–376. doi: 10.1128/mr.53.3.367-376.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledebur H., Nixon B. T. Tandem DctD-binding sites of the Rhizobium meliloti dctA upstream activating sequence are essential for optimal function despite a 50- to 100-fold difference in affinity for DctD. Mol Microbiol. 1992 Dec;6(23):3479–3492. doi: 10.1111/j.1365-2958.1992.tb01783.x. [DOI] [PubMed] [Google Scholar]
- Magasanik B. Reversible phosphorylation of an enhancer binding protein regulates the transcription of bacterial nitrogen utilization genes. Trends Biochem Sci. 1988 Dec;13(12):475–479. doi: 10.1016/0968-0004(88)90234-4. [DOI] [PubMed] [Google Scholar]
- Markwell M. A., Haas S. M., Bieber L. L., Tolbert N. E. A modification of the Lowry procedure to simplify protein determination in membrane and lipoprotein samples. Anal Biochem. 1978 Jun 15;87(1):206–210. doi: 10.1016/0003-2697(78)90586-9. [DOI] [PubMed] [Google Scholar]
- Masepohl B., Klipp W., Pühler A. Genetic characterization and sequence analysis of the duplicated nifA/nifB gene region of Rhodobacter capsulatus. Mol Gen Genet. 1988 Apr;212(1):27–37. doi: 10.1007/BF00322441. [DOI] [PubMed] [Google Scholar]
- Maupin J. A., Shanmugam K. T. Genetic regulation of formate hydrogenlyase of Escherichia coli: role of the fhlA gene product as a transcriptional activator for a new regulatory gene, fhlB. J Bacteriol. 1990 Sep;172(9):4798–4806. doi: 10.1128/jb.172.9.4798-4806.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrick M. J. In a class of its own--the RNA polymerase sigma factor sigma 54 (sigma N). Mol Microbiol. 1993 Dec;10(5):903–909. doi: 10.1111/j.1365-2958.1993.tb00961.x. [DOI] [PubMed] [Google Scholar]
- Morett E., Buck M. In vivo studies on the interaction of RNA polymerase-sigma 54 with the Klebsiella pneumoniae and Rhizobium meliloti nifH promoters. The role of NifA in the formation of an open promoter complex. J Mol Biol. 1989 Nov 5;210(1):65–77. doi: 10.1016/0022-2836(89)90291-x. [DOI] [PubMed] [Google Scholar]
- Morett E., Buck M. NifA-dependent in vivo protection demonstrates that the upstream activator sequence of nif promoters is a protein binding site. Proc Natl Acad Sci U S A. 1988 Dec;85(24):9401–9405. doi: 10.1073/pnas.85.24.9401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morett E., Segovia L. The sigma 54 bacterial enhancer-binding protein family: mechanism of action and phylogenetic relationship of their functional domains. J Bacteriol. 1993 Oct;175(19):6067–6074. doi: 10.1128/jb.175.19.6067-6074.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ninfa A. J., Magasanik B. Covalent modification of the glnG product, NRI, by the glnL product, NRII, regulates the transcription of the glnALG operon in Escherichia coli. Proc Natl Acad Sci U S A. 1986 Aug;83(16):5909–5913. doi: 10.1073/pnas.83.16.5909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ninfa A. J., Reitzer L. J., Magasanik B. Initiation of transcription at the bacterial glnAp2 promoter by purified E. coli components is facilitated by enhancers. Cell. 1987 Sep 25;50(7):1039–1046. doi: 10.1016/0092-8674(87)90170-x. [DOI] [PubMed] [Google Scholar]
- North A. K., Klose K. E., Stedman K. M., Kustu S. Prokaryotic enhancer-binding proteins reflect eukaryote-like modularity: the puzzle of nitrogen regulatory protein C. J Bacteriol. 1993 Jul;175(14):4267–4273. doi: 10.1128/jb.175.14.4267-4273.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson J. S. Signal transduction schemes of bacteria. Cell. 1993 Jun 4;73(5):857–871. doi: 10.1016/0092-8674(93)90267-t. [DOI] [PubMed] [Google Scholar]
- Pittard A. J., Davidson B. E. TyrR protein of Escherichia coli and its role as repressor and activator. Mol Microbiol. 1991 Jul;5(7):1585–1592. doi: 10.1111/j.1365-2958.1991.tb01904.x. [DOI] [PubMed] [Google Scholar]
- Popham D. L., Szeto D., Keener J., Kustu S. Function of a bacterial activator protein that binds to transcriptional enhancers. Science. 1989 Feb 3;243(4891):629–635. doi: 10.1126/science.2563595. [DOI] [PubMed] [Google Scholar]
- Porter S. C., North A. K., Wedel A. B., Kustu S. Oligomerization of NTRC at the glnA enhancer is required for transcriptional activation. Genes Dev. 1993 Nov;7(11):2258–2273. doi: 10.1101/gad.7.11.2258. [DOI] [PubMed] [Google Scholar]
- Preker P., Hübner P., Schmehl M., Klipp W., Bickle T. A. Mapping and characterization of the promoter elements of the regulatory nif genes rpoN, nifA1 and nifA2 in Rhodobacter capsulatus. Mol Microbiol. 1992 Apr;6(8):1035–1047. doi: 10.1111/j.1365-2958.1992.tb02169.x. [DOI] [PubMed] [Google Scholar]
- Ramakrishnan G., Newton A. FlbD of Caulobacter crescentus is a homologue of the NtrC (NRI) protein and activates sigma 54-dependent flagellar gene promoters. Proc Natl Acad Sci U S A. 1990 Mar;87(6):2369–2373. doi: 10.1073/pnas.87.6.2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reitzer L. J., Magasanik B. Isolation of the nitrogen assimilation regulator NR(I), the product of the glnG gene of Escherichia coli. Proc Natl Acad Sci U S A. 1983 Sep;80(18):5554–5558. doi: 10.1073/pnas.80.18.5554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reitzer L. J., Magasanik B. Transcription of glnA in E. coli is stimulated by activator bound to sites far from the promoter. Cell. 1986 Jun 20;45(6):785–792. doi: 10.1016/0092-8674(86)90553-2. [DOI] [PubMed] [Google Scholar]
- Ronson C. W., Astwood P. M., Nixon B. T., Ausubel F. M. Deduced products of C4-dicarboxylate transport regulatory genes of Rhizobium leguminosarum are homologous to nitrogen regulatory gene products. Nucleic Acids Res. 1987 Oct 12;15(19):7921–7934. doi: 10.1093/nar/15.19.7921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santero E., Hoover T. R., North A. K., Berger D. K., Porter S. C., Kustu S. Role of integration host factor in stimulating transcription from the sigma 54-dependent nifH promoter. J Mol Biol. 1992 Oct 5;227(3):602–620. doi: 10.1016/0022-2836(92)90211-2. [DOI] [PubMed] [Google Scholar]
- Schlensog V., Böck A. Identification and sequence analysis of the gene encoding the transcriptional activator of the formate hydrogenlyase system of Escherichia coli. Mol Microbiol. 1990 Aug;4(8):1319–1327. doi: 10.1111/j.1365-2958.1990.tb00711.x. [DOI] [PubMed] [Google Scholar]
- Stock J. B., Ninfa A. J., Stock A. M. Protein phosphorylation and regulation of adaptive responses in bacteria. Microbiol Rev. 1989 Dec;53(4):450–490. doi: 10.1128/mr.53.4.450-490.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studier F. W., Moffatt B. A. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol. 1986 May 5;189(1):113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- Su W., Porter S., Kustu S., Echols H. DNA-looping and enhancer activity: association between DNA-bound NtrC activator and RNA polymerase at the bacterial glnA promoter. Proc Natl Acad Sci U S A. 1990 Jul;87(14):5504–5508. doi: 10.1073/pnas.87.14.5504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira J., Messing J. Production of single-stranded plasmid DNA. Methods Enzymol. 1987;153:3–11. doi: 10.1016/0076-6879(87)53044-0. [DOI] [PubMed] [Google Scholar]
- Volz K. Structural conservation in the CheY superfamily. Biochemistry. 1993 Nov 9;32(44):11741–11753. doi: 10.1021/bi00095a001. [DOI] [PubMed] [Google Scholar]
- Walker J. E., Saraste M., Runswick M. J., Gay N. J. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1(8):945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall J. D., Braddock K. Mapping of Rhodopseudomonas capsulata nif genes. J Bacteriol. 1984 May;158(2):404–410. doi: 10.1128/jb.158.2.404-410.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wedel A., Weiss D. S., Popham D., Dröge P., Kustu S. A bacterial enhancer functions to tether a transcriptional activator near a promoter. Science. 1990 Apr 27;248(4954):486–490. doi: 10.1126/science.1970441. [DOI] [PubMed] [Google Scholar]
- Weiss D. S., Batut J., Klose K. E., Keener J., Kustu S. The phosphorylated form of the enhancer-binding protein NTRC has an ATPase activity that is essential for activation of transcription. Cell. 1991 Oct 4;67(1):155–167. doi: 10.1016/0092-8674(91)90579-n. [DOI] [PubMed] [Google Scholar]
- Weiss V., Claverie-Martin F., Magasanik B. Phosphorylation of nitrogen regulator I of Escherichia coli induces strong cooperative binding to DNA essential for activation of transcription. Proc Natl Acad Sci U S A. 1992 Jun 1;89(11):5088–5092. doi: 10.1073/pnas.89.11.5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehall S., Austin S., Dixon R. DNA supercoiling response of the sigma 54-dependent Klebsiella pneumoniae nifL promoter in vitro. J Mol Biol. 1992 Jun 5;225(3):591–607. doi: 10.1016/0022-2836(92)90388-z. [DOI] [PubMed] [Google Scholar]
- Wong P. K., Popham D., Keener J., Kustu S. In vitro transcription of the nitrogen fixation regulatory operon nifLA of Klebsiella pneumoniae. J Bacteriol. 1987 Jun;169(6):2876–2880. doi: 10.1128/jb.169.6.2876-2880.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Ganesan S., Sarsero J., Pittard A. J. A genetic analysis of various functions of the TyrR protein of Escherichia coli. J Bacteriol. 1993 Mar;175(6):1767–1776. doi: 10.1128/jb.175.6.1767-1776.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen H. C., Marrs B. Map of genes for carotenoid and bacteriochlorophyll biosynthesis in Rhodopseudomonas capsulata. J Bacteriol. 1976 May;126(2):619–629. doi: 10.1128/jb.126.2.619-629.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lorenzo V., Herrero M., Metzke M., Timmis K. N. An upstream XylR- and IHF-induced nucleoprotein complex regulates the sigma 54-dependent Pu promoter of TOL plasmid. EMBO J. 1991 May;10(5):1159–1167. doi: 10.1002/j.1460-2075.1991.tb08056.x. [DOI] [PMC free article] [PubMed] [Google Scholar]