Abstract
DNA polymerase II (Pol II) is regulated as part of the SOS response to DNA damage in Escherichia coli. We examined the participation of Pol II in the response to oxidative damage, adaptive mutation, and recombination. Cells lacking Pol II activity (polB delta 1 mutants) exhibited 5- to 10-fold-greater sensitivity to mode 1 killing by H2O2 compared with isogenic polB+ cells. Survival decreased by about 15-fold when polB mutants containing defective superoxide dismutase genes, sodA and sodB, were compared with polB+ sodA sodB mutants. Resistance to peroxide killing was restored following P1 transduction of polB cells to polB+ or by conjugation of polB cells with an F' plasmid carrying a copy of polB+. The rate at which Lac+ mutations arose in Lac- cells subjected to selection for lactose utilization, a phenomenon known as adaptive mutation, was increased threefold in polB backgrounds and returned to wild-type rates when polB cells were transduced to polB+. Following multiple passages of polB cells or prolonged starvation, a progressive loss of sensitivity to killing by peroxide was observed, suggesting that second-site suppressor mutations may be occurring with relatively high frequencies. The presence of suppressor mutations may account for the apparent lack of a mutant phenotype in earlier studies. A well-established polB strain, a dinA Mu d(Apr lac) fusion (GW1010), exhibited wild-type (Pol II+) sensitivity to killing by peroxide, consistent with the accumulation of second-site suppressor mutations. A high titer anti-Pol II polyclonal antibody was used to screen for the presence of Pol II in other bacteria and in the yeast Saccharomyces cerevisiae. Cross-reacting material was found in all gram-negative strains tested but was not detected in gram-positive strains or in S. cerevisiae. Induction of Pol II by nalidixic acid was observed in E. coli K-12, B, and C, in Shigella flexneri, and in Salmonella typhimurium.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Beauchamp C., Fridovich I. Superoxide dismutase: improved assays and an assay applicable to acrylamide gels. Anal Biochem. 1971 Nov;44(1):276–287. doi: 10.1016/0003-2697(71)90370-8. [DOI] [PubMed] [Google Scholar]
- Bonner C. A., Hays S., McEntee K., Goodman M. F. DNA polymerase II is encoded by the DNA damage-inducible dinA gene of Escherichia coli. Proc Natl Acad Sci U S A. 1990 Oct;87(19):7663–7667. doi: 10.1073/pnas.87.19.7663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonner C. A., Randall S. K., Rayssiguier C., Radman M., Eritja R., Kaplan B. E., McEntee K., Goodman M. F. Purification and characterization of an inducible Escherichia coli DNA polymerase capable of insertion and bypass at abasic lesions in DNA. J Biol Chem. 1988 Dec 15;263(35):18946–18952. [PubMed] [Google Scholar]
- Bonner C. A., Stukenberg P. T., Rajagopalan M., Eritja R., O'Donnell M., McEntee K., Echols H., Goodman M. F. Processive DNA synthesis by DNA polymerase II mediated by DNA polymerase III accessory proteins. J Biol Chem. 1992 Jun 5;267(16):11431–11438. [PubMed] [Google Scholar]
- Cairns J., Foster P. L. Adaptive reversion of a frameshift mutation in Escherichia coli. Genetics. 1991 Aug;128(4):695–701. doi: 10.1093/genetics/128.4.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calos M. P., Miller J. H. Genetic and sequence analysis of frameshift mutations induced by ICR-191. J Mol Biol. 1981 Nov 25;153(1):39–64. doi: 10.1016/0022-2836(81)90525-8. [DOI] [PubMed] [Google Scholar]
- Campbell J. L., Soll L., Richardson C. C. Isolation and partial characterization of a mutant of Escherichia coli deficient in DNA polymerase II. Proc Natl Acad Sci U S A. 1972 Aug;69(8):2090–2094. doi: 10.1073/pnas.69.8.2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadaban M. J., Cohen S. N. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J Mol Biol. 1980 Apr;138(2):179–207. doi: 10.1016/0022-2836(80)90283-1. [DOI] [PubMed] [Google Scholar]
- Chen H., Bryan S. K., Moses R. E. Cloning the polB gene of Escherichia coli and identification of its product. J Biol Chem. 1989 Dec 5;264(34):20591–20595. [PubMed] [Google Scholar]
- Chen H., Sun Y., Stark T., Beattie W., Moses R. E. Nucleotide sequence and deletion analysis of the polB gene of Escherichia coli. DNA Cell Biol. 1990 Nov;9(9):631–635. doi: 10.1089/dna.1990.9.631. [DOI] [PubMed] [Google Scholar]
- De Lucia P., Cairns J. Isolation of an E. coli strain with a mutation affecting DNA polymerase. Nature. 1969 Dec 20;224(5225):1164–1166. doi: 10.1038/2241164a0. [DOI] [PubMed] [Google Scholar]
- DiFrancesco R., Bhatnagar S. K., Brown A., Bessman M. J. The interaction of DNA polymerase III and the product of the Escherichia coli mutator gene, mutD. J Biol Chem. 1984 May 10;259(9):5567–5573. [PubMed] [Google Scholar]
- Foster P. L., Cairns J. Mechanisms of directed mutation. Genetics. 1992 Aug;131(4):783–789. doi: 10.1093/genetics/131.4.783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster P. L., Trimarchi J. M. Adaptive reversion of a frameshift mutation in Escherichia coli by simple base deletions in homopolymeric runs. Science. 1994 Jul 15;265(5170):407–409. doi: 10.1126/science.8023164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gefter M. L., Hirota Y., Kornberg T., Wechsler J. A., Barnoux C. Analysis of DNA polymerases II and 3 in mutants of Escherichia coli thermosensitive for DNA synthesis. Proc Natl Acad Sci U S A. 1971 Dec;68(12):3150–3153. doi: 10.1073/pnas.68.12.3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goerlich O., Quillardet P., Hofnung M. Induction of the SOS response by hydrogen peroxide in various Escherichia coli mutants with altered protection against oxidative DNA damage. J Bacteriol. 1989 Nov;171(11):6141–6147. doi: 10.1128/jb.171.11.6141-6147.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris R. S., Longerich S., Rosenberg S. M. Recombination in adaptive mutation. Science. 1994 Apr 8;264(5156):258–260. doi: 10.1126/science.8146657. [DOI] [PubMed] [Google Scholar]
- Horii Z., Clark A. J. Genetic analysis of the recF pathway to genetic recombination in Escherichia coli K12: isolation and characterization of mutants. J Mol Biol. 1973 Oct 25;80(2):327–344. doi: 10.1016/0022-2836(73)90176-9. [DOI] [PubMed] [Google Scholar]
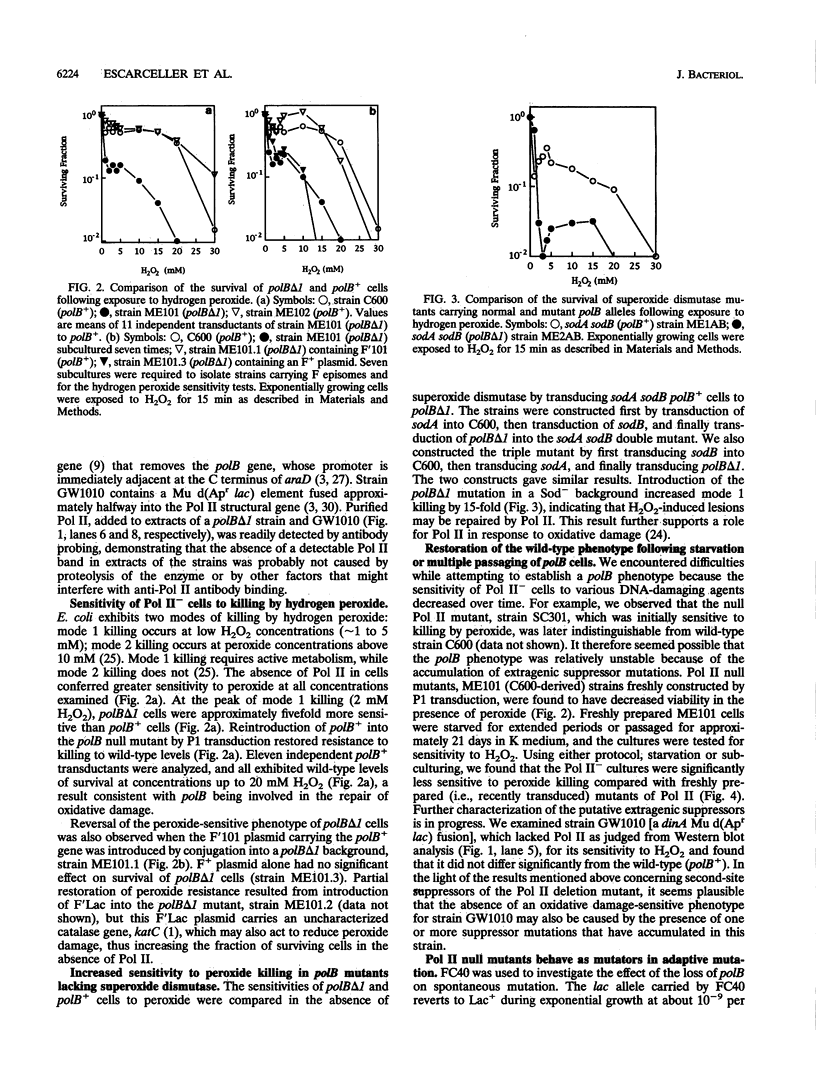
- Hughes A. J., Jr, Bryan S. K., Chen H., Moses R. E., McHenry C. S. Escherichia coli DNA polymerase II is stimulated by DNA polymerase III holoenzyme auxiliary subunits. J Biol Chem. 1991 Mar 5;266(7):4568–4573. [PubMed] [Google Scholar]
- Imlay J. A., Linn S. Bimodal pattern of killing of DNA-repair-defective or anoxically grown Escherichia coli by hydrogen peroxide. J Bacteriol. 1986 May;166(2):519–527. doi: 10.1128/jb.166.2.519-527.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imlay J. A., Linn S. DNA damage and oxygen radical toxicity. Science. 1988 Jun 3;240(4857):1302–1309. doi: 10.1126/science.3287616. [DOI] [PubMed] [Google Scholar]
- Imlay J. A., Linn S. Mutagenesis and stress responses induced in Escherichia coli by hydrogen peroxide. J Bacteriol. 1987 Jul;169(7):2967–2976. doi: 10.1128/jb.169.7.2967-2976.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
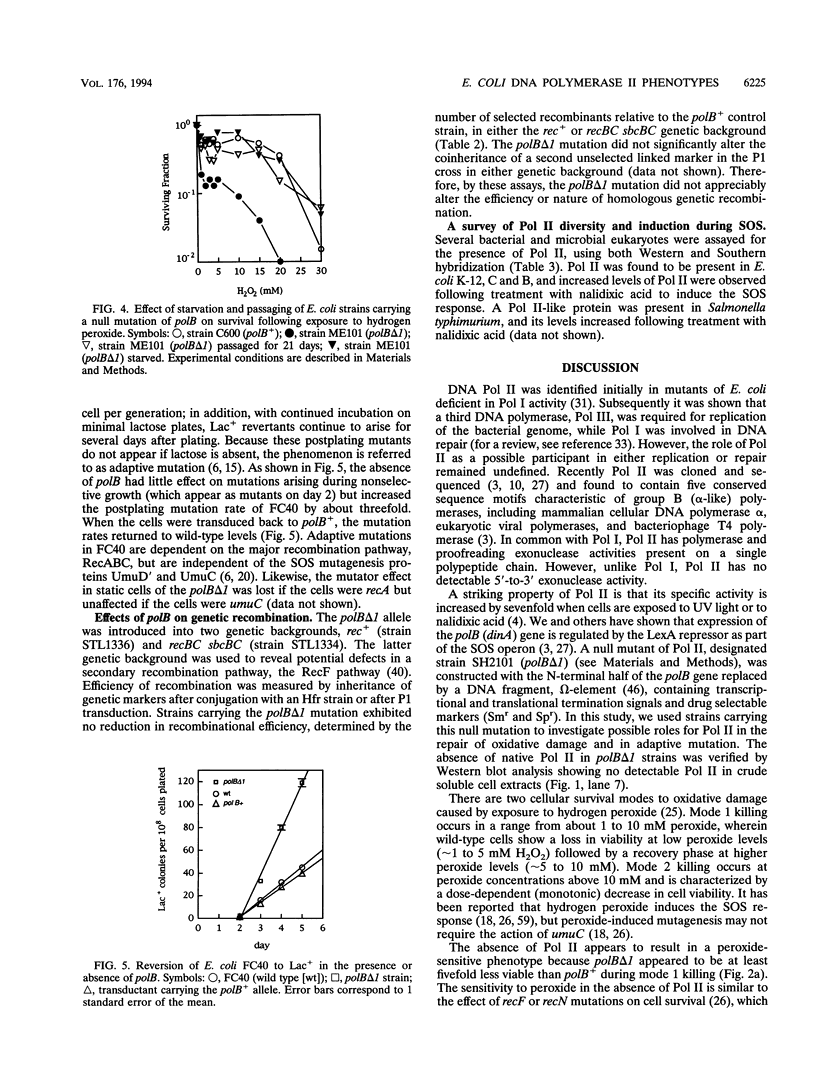
- Iwasaki H., Nakata A., Walker G. C., Shinagawa H. The Escherichia coli polB gene, which encodes DNA polymerase II, is regulated by the SOS system. J Bacteriol. 1990 Nov;172(11):6268–6273. doi: 10.1128/jb.172.11.6268-6273.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins D. E., Schultz J. E., Matin A. Starvation-induced cross protection against heat or H2O2 challenge in Escherichia coli. J Bacteriol. 1988 Sep;170(9):3910–3914. doi: 10.1128/jb.170.9.3910-3914.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knippers R. DNA polymerase II. Nature. 1970 Dec 12;228(5276):1050–1053. doi: 10.1038/2281050a0. [DOI] [PubMed] [Google Scholar]
- Kohara Y., Akiyama K., Isono K. The physical map of the whole E. coli chromosome: application of a new strategy for rapid analysis and sorting of a large genomic library. Cell. 1987 Jul 31;50(3):495–508. doi: 10.1016/0092-8674(87)90503-4. [DOI] [PubMed] [Google Scholar]
- Kornberg T., Gefter M. L. DNA synthesis in cell-free extracts of a DNA polymerase-defective mutant. Biochem Biophys Res Commun. 1970 Sep 30;40(6):1348–1355. doi: 10.1016/0006-291x(70)90014-8. [DOI] [PubMed] [Google Scholar]
- Kornberg T., Gefter M. L. Purification and DNA synthesis in cell-free extracts: properties of DNA polymerase II. Proc Natl Acad Sci U S A. 1971 Apr;68(4):761–764. doi: 10.1073/pnas.68.4.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Low B. Formation of merodiploids in matings with a class of Rec- recipient strains of Escherichia coli K12. Proc Natl Acad Sci U S A. 1968 May;60(1):160–167. doi: 10.1073/pnas.60.1.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low R. L., Rashbaum S. A., Cozzarelli N. R. Purification and characterization of DNA polymerase III from Bacillus subtilis. J Biol Chem. 1976 Mar 10;251(5):1311–1325. [PubMed] [Google Scholar]
- Marshall T., Williams K. M. Centriprep ultrafiltration for fractionation of serum and urinary proteins before electrophoresis. Clin Chem. 1993 Jul;39(7):1558–1558. [PubMed] [Google Scholar]
- McCann M. P., Kidwell J. P., Matin A. The putative sigma factor KatF has a central role in development of starvation-mediated general resistance in Escherichia coli. J Bacteriol. 1991 Jul;173(13):4188–4194. doi: 10.1128/jb.173.13.4188-4194.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHenry C. S. DNA polymerase III holoenzyme of Escherichia coli. Annu Rev Biochem. 1988;57:519–550. doi: 10.1146/annurev.bi.57.070188.002511. [DOI] [PubMed] [Google Scholar]
- Moses R. E., Richardson C. C. A new DNA polymerase activity of Escherichia coli. I. Purification and properties of the activity present in E. coli polA1. Biochem Biophys Res Commun. 1970 Dec 24;41(6):1557–1564. doi: 10.1016/0006-291x(70)90565-6. [DOI] [PubMed] [Google Scholar]
- Prentki P., Krisch H. M. In vitro insertional mutagenesis with a selectable DNA fragment. Gene. 1984 Sep;29(3):303–313. doi: 10.1016/0378-1119(84)90059-3. [DOI] [PubMed] [Google Scholar]
- Rosenberg S. M., Longerich S., Gee P., Harris R. S. Adaptive mutation by deletions in small mononucleotide repeats. Science. 1994 Jul 15;265(5170):405–407. doi: 10.1126/science.8023163. [DOI] [PubMed] [Google Scholar]
- Schaaper R. M. Mechanisms of mutagenesis in the Escherichia coli mutator mutD5: role of DNA mismatch repair. Proc Natl Acad Sci U S A. 1988 Nov;85(21):8126–8130. doi: 10.1073/pnas.85.21.8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaaper R. M., Radman M. The extreme mutator effect of Escherichia coli mutD5 results from saturation of mismatch repair by excessive DNA replication errors. EMBO J. 1989 Nov;8(11):3511–3516. doi: 10.1002/j.1460-2075.1989.tb08516.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheuermann R. H., Echols H. A separate editing exonuclease for DNA replication: the epsilon subunit of Escherichia coli DNA polymerase III holoenzyme. Proc Natl Acad Sci U S A. 1984 Dec;81(24):7747–7751. doi: 10.1073/pnas.81.24.7747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheuermann R., Tam S., Burgers P. M., Lu C., Echols H. Identification of the epsilon-subunit of Escherichia coli DNA polymerase III holoenzyme as the dnaQ gene product: a fidelity subunit for DNA replication. Proc Natl Acad Sci U S A. 1983 Dec;80(23):7085–7089. doi: 10.1073/pnas.80.23.7085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer M., Baker T. A., Schnitzler G., Deischel S. M., Goel M., Dove W., Jaacks K. J., Grossman A. D., Erickson J. W., Gross C. A. A collection of strains containing genetically linked alternating antibiotic resistance elements for genetic mapping of Escherichia coli. Microbiol Rev. 1989 Mar;53(1):1–24. doi: 10.1128/mr.53.1.1-24.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternberg N. L., Maurer R. Bacteriophage-mediated generalized transduction in Escherichia coli and Salmonella typhimurium. Methods Enzymol. 1991;204:18–43. doi: 10.1016/0076-6879(91)04004-8. [DOI] [PubMed] [Google Scholar]
- Tessman I., Kennedy M. A. DNA polymerase II of Escherichia coli in the bypass of abasic sites in vivo. Genetics. 1994 Feb;136(2):439–448. doi: 10.1093/genetics/136.2.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas N., Jones C. N., Thomas P. L. Low volume processing of protein blots in rolling drums. Anal Biochem. 1988 May 1;170(2):393–396. doi: 10.1016/0003-2697(88)90650-1. [DOI] [PubMed] [Google Scholar]
- Walker G. C. Mutagenesis and inducible responses to deoxyribonucleic acid damage in Escherichia coli. Microbiol Rev. 1984 Mar;48(1):60–93. doi: 10.1128/mr.48.1.60-93.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanner B. L. Novel regulatory mutants of the phosphate regulon in Escherichia coli K-12. J Mol Biol. 1986 Sep 5;191(1):39–58. doi: 10.1016/0022-2836(86)90421-3. [DOI] [PubMed] [Google Scholar]
- Wickner S., Hurwitz J. Conversion of phiX174 viral DNA to double-stranded form by purified Escherichia coli proteins. Proc Natl Acad Sci U S A. 1974 Oct;71(10):4120–4124. doi: 10.1073/pnas.71.10.4120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willetts N. S., Clark A. J., Low B. Genetic location of certain mutations conferring recombination deficiency in Escherichia coli. J Bacteriol. 1969 Jan;97(1):244–249. doi: 10.1128/jb.97.1.244-249.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese C. R. Bacterial evolution. Microbiol Rev. 1987 Jun;51(2):221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]



