Abstract
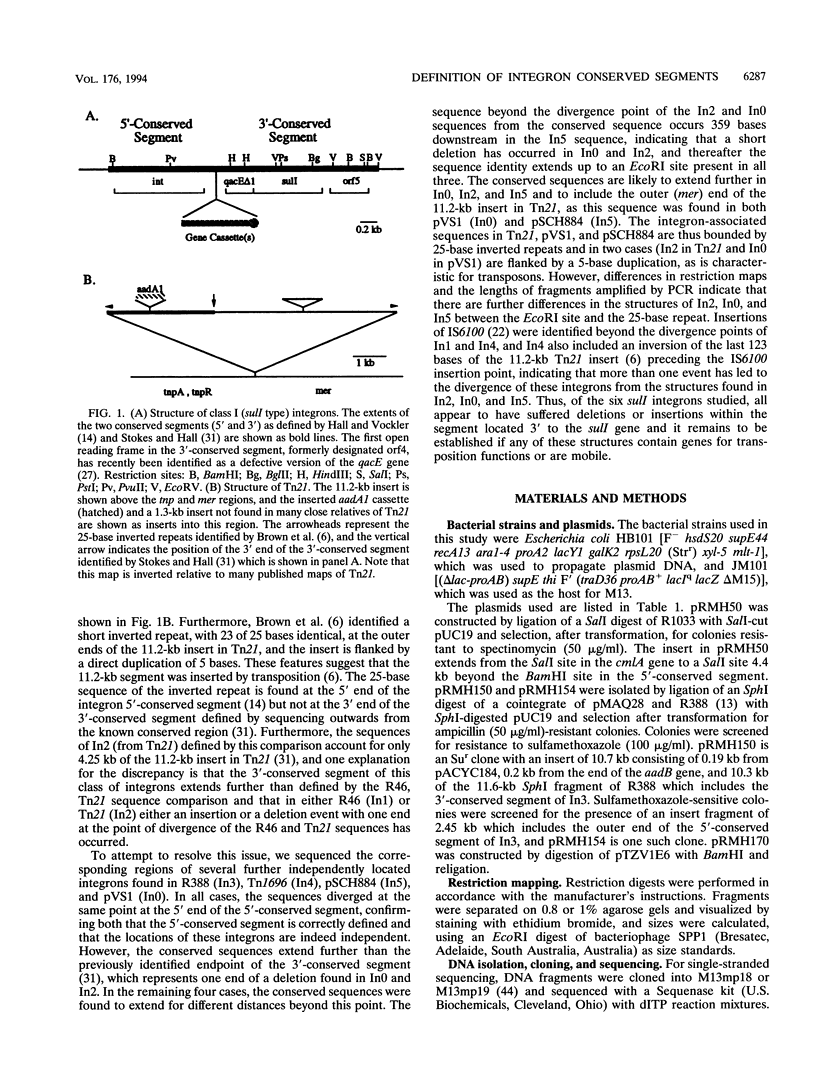
The positions of the outer boundaries of the 5'- and 3'-conserved segment sequences of integrons found at several different locations have been determined. The position of the 5' end of the 5'-conserved segment is the same for six independently located integrons, In1 (R46), In2 (Tn21), In3 (R388), In4 (Tn1696), In5 (pSCH884), and In0 (pVS1). However, the extent of the 3'-conserved segment differs in each integron. The sequences of In2 and In0 diverge first from the conserved sequence, and their divergence point corresponds to the 3'-conserved segment endpoint defined previously (H.W. Stokes and R.M. Hall, Mol. Microbiol. 3:1669-1683, 1989), which now represents the endpoint of a 359-base deletion in In0 and In2. The sequence identity in In3, In1, In4, and In5 extends beyond this point, but each sequence diverges from the conserved sequence at a different point within a short region. Insertions of IS6100 were identified adjacent to the end of the conserved region in In1 and 123 bases beyond the divergence point of In4. These 123 bases are identical to the sequence found at the mer end of the 11.2-kb insertion in Tn21 but are inverted. In5 and In0 are bounded by the same 25-base inverted repeat that bounds the 11.2-kb insert in Tn21, and this insert now corresponds to In2. However, while In0, In2, and In5 have features characteristic of transposable elements, differences in the structures of these three integrons and the absence of evidence of mobility currently preclude the identification of all of the sequences associated with a functional transposon of this type.
Full text
PDF







Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Avila P., de la Cruz F. Physical and genetic map of the IncW plasmid R388. Plasmid. 1988 Sep;20(2):155–157. doi: 10.1016/0147-619x(88)90019-4. [DOI] [PubMed] [Google Scholar]
- Bissonnette L., Champetier S., Buisson J. P., Roy P. H. Characterization of the nonenzymatic chloramphenicol resistance (cmlA) gene of the In4 integron of Tn1696: similarity of the product to transmembrane transport proteins. J Bacteriol. 1991 Jul;173(14):4493–4502. doi: 10.1128/jb.173.14.4493-4502.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bissonnette L., Roy P. H. Characterization of In0 of Pseudomonas aeruginosa plasmid pVS1, an ancestor of integrons of multiresistance plasmids and transposons of gram-negative bacteria. J Bacteriol. 1992 Feb;174(4):1248–1257. doi: 10.1128/jb.174.4.1248-1257.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown A. M., Coupland G. M., Willetts N. S. Characterization of IS46, an insertion sequence found on two IncN plasmids. J Bacteriol. 1984 Aug;159(2):472–481. doi: 10.1128/jb.159.2.472-481.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown A. M., Willetts N. S. A physical and genetic map of the IncN plasmid R46. Plasmid. 1981 Mar;5(2):188–201. doi: 10.1016/0147-619x(81)90020-2. [DOI] [PubMed] [Google Scholar]
- Brown N. L., Misra T. K., Winnie J. N., Schmidt A., Seiff M., Silver S. The nucleotide sequence of the mercuric resistance operons of plasmid R100 and transposon Tn501: further evidence for mer genes which enhance the activity of the mercuric ion detoxification system. Mol Gen Genet. 1986 Jan;202(1):143–151. doi: 10.1007/BF00330531. [DOI] [PubMed] [Google Scholar]
- Cameron F. H., Groot Obbink D. J., Ackerman V. P., Hall R. M. Nucleotide sequence of the AAD(2'') aminoglycoside adenylyltransferase determinant aadB. Evolutionary relationship of this region with those surrounding aadA in R538-1 and dhfrII in R388. Nucleic Acids Res. 1986 Nov 11;14(21):8625–8635. doi: 10.1093/nar/14.21.8625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collis C. M., Grammaticopoulos G., Briton J., Stokes H. W., Hall R. M. Site-specific insertion of gene cassettes into integrons. Mol Microbiol. 1993 Jul;9(1):41–52. doi: 10.1111/j.1365-2958.1993.tb01667.x. [DOI] [PubMed] [Google Scholar]
- Collis C. M., Hall R. M. Gene cassettes from the insert region of integrons are excised as covalently closed circles. Mol Microbiol. 1992 Oct;6(19):2875–2885. doi: 10.1111/j.1365-2958.1992.tb01467.x. [DOI] [PubMed] [Google Scholar]
- Collis C. M., Hall R. M. Site-specific deletion and rearrangement of integron insert genes catalyzed by the integron DNA integrase. J Bacteriol. 1992 Mar;174(5):1574–1585. doi: 10.1128/jb.174.5.1574-1585.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diver W. P., Grinsted J., Fritzinger D. C., Brown N. L., Altenbuchner J., Rogowsky P., Schmitt R. DNA sequences of and complementation by the tnpR genes of Tn21, Tn501 and Tn1721. Mol Gen Genet. 1983;191(2):189–193. doi: 10.1007/BF00334812. [DOI] [PubMed] [Google Scholar]
- Hall R. M., Brookes D. E., Stokes H. W. Site-specific insertion of genes into integrons: role of the 59-base element and determination of the recombination cross-over point. Mol Microbiol. 1991 Aug;5(8):1941–1959. doi: 10.1111/j.1365-2958.1991.tb00817.x. [DOI] [PubMed] [Google Scholar]
- Hall R. M., Vockler C. The region of the IncN plasmid R46 coding for resistance to beta-lactam antibiotics, streptomycin/spectinomycin and sulphonamides is closely related to antibiotic resistance segments found in IncW plasmids and in Tn21-like transposons. Nucleic Acids Res. 1987 Sep 25;15(18):7491–7501. doi: 10.1093/nar/15.18.7491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch P. R., Wang C. L., Woodward M. J. Construction of a Tn5 derivative determining resistance to gentamicin and spectinomycin using a fragment cloned from R1033. Gene. 1986;48(2-3):203–209. doi: 10.1016/0378-1119(86)90078-8. [DOI] [PubMed] [Google Scholar]
- Hollingshead S., Vapnek D. Nucleotide sequence analysis of a gene encoding a streptomycin/spectinomycin adenylyltransferase. Plasmid. 1985 Jan;13(1):17–30. doi: 10.1016/0147-619x(85)90052-6. [DOI] [PubMed] [Google Scholar]
- Hsiao K. A fast and simple procedure for sequencing double stranded DNA with sequenase. Nucleic Acids Res. 1991 May 25;19(10):2787–2787. doi: 10.1093/nar/19.10.2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyde D. R., Tu C. P. tnpM: a novel regulatory gene that enhances Tn21 transposition and suppresses cointegrate resolution. Cell. 1985 Sep;42(2):629–638. doi: 10.1016/0092-8674(85)90120-5. [DOI] [PubMed] [Google Scholar]
- Itoh Y., Watson J. M., Haas D., Leisinger T. Genetic and molecular characterization of the Pseudomonas plasmid pVS1. Plasmid. 1984 May;11(3):206–220. doi: 10.1016/0147-619x(84)90027-1. [DOI] [PubMed] [Google Scholar]
- Martin C., Timm J., Rauzier J., Gomez-Lus R., Davies J., Gicquel B. Transposition of an antibiotic resistance element in mycobacteria. Nature. 1990 Jun 21;345(6277):739–743. doi: 10.1038/345739a0. [DOI] [PubMed] [Google Scholar]
- Mercier J., Lachapelle J., Couture F., Lafond M., Vézina G., Boissinot M., Levesque R. C. Structural and functional characterization of tnpI, a recombinase locus in Tn21 and related beta-lactamase transposons. J Bacteriol. 1990 Jul;172(7):3745–3757. doi: 10.1128/jb.172.7.3745-3757.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer J. F., Nies B. A., Kratz J., Wiedemann B. Evolution of Tn21-related transposons: isolation of Tn2425, which harbours IS161. J Gen Microbiol. 1985 May;131(5):1123–1130. doi: 10.1099/00221287-131-5-1123. [DOI] [PubMed] [Google Scholar]
- Ouellette M., Roy P. H. Homology of ORFs from Tn2603 and from R46 to site-specific recombinases. Nucleic Acids Res. 1987 Dec 10;15(23):10055–10055. doi: 10.1093/nar/15.23.10055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parent R., Roy P. H. The chloramphenicol acetyltransferase gene of Tn2424: a new breed of cat. J Bacteriol. 1992 May;174(9):2891–2897. doi: 10.1128/jb.174.9.2891-2897.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsen I. T., Littlejohn T. G., Rådström P., Sundström L., Sköld O., Swedberg G., Skurray R. A. The 3' conserved segment of integrons contains a gene associated with multidrug resistance to antiseptics and disinfectants. Antimicrob Agents Chemother. 1993 Apr;37(4):761–768. doi: 10.1128/aac.37.4.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt F., Klopfer-Kaul I. Evolutionary relationship between Tn21-like elements and pBP201, a plasmid from Klebsiella pneumoniae mediating resistance to gentamicin and eight other drugs. Mol Gen Genet. 1984;197(1):109–119. doi: 10.1007/BF00327930. [DOI] [PubMed] [Google Scholar]
- Shaw K. J., Cramer C. A., Rizzo M., Mierzwa R., Gewain K., Miller G. H., Hare R. S. Isolation, characterization, and DNA sequence analysis of an AAC(6')-II gene from Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1989 Dec;33(12):2052–2062. doi: 10.1128/aac.33.12.2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinclair M. I., Holloway B. W. A chromosomally located transposon in Pseudomonas aeruginosa. J Bacteriol. 1982 Aug;151(2):569–579. doi: 10.1128/jb.151.2.569-579.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephen D., Jones C., Schofield J. P. A rapid method for isolating high quality plasmid DNA suitable for DNA sequencing. Nucleic Acids Res. 1990 Dec 25;18(24):7463–7464. doi: 10.1093/nar/18.24.7463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokes H. W., Hall R. M. A novel family of potentially mobile DNA elements encoding site-specific gene-integration functions: integrons. Mol Microbiol. 1989 Dec;3(12):1669–1683. doi: 10.1111/j.1365-2958.1989.tb00153.x. [DOI] [PubMed] [Google Scholar]
- Stokes H. W., Hall R. M. Sequence analysis of the inducible chloramphenicol resistance determinant in the Tn1696 integron suggests regulation by translational attenuation. Plasmid. 1991 Jul;26(1):10–19. doi: 10.1016/0147-619x(91)90032-r. [DOI] [PubMed] [Google Scholar]
- Stokes H. W., Hall R. M. The integron In1 in plasmid R46 includes two copies of the oxa2 gene cassette. Plasmid. 1992 Nov;28(3):225–234. doi: 10.1016/0147-619x(92)90054-e. [DOI] [PubMed] [Google Scholar]
- Sundström L., Rådström P., Swedberg G., Sköld O. Site-specific recombination promotes linkage between trimethoprim- and sulfonamide resistance genes. Sequence characterization of dhfrV and sulI and a recombination active locus of Tn21. Mol Gen Genet. 1988 Aug;213(2-3):191–201. doi: 10.1007/BF00339581. [DOI] [PubMed] [Google Scholar]
- Sundström L., Swedberg G., Sköld O. Characterization of transposon Tn5086, carrying the site-specifically inserted gene dhfrVII mediating trimethoprim resistance. J Bacteriol. 1993 Mar;175(6):1796–1805. doi: 10.1128/jb.175.6.1796-1805.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundström L., Vinayagamoorthy T., Sköld O. Novel type of plasmid-borne resistance to trimethoprim. Antimicrob Agents Chemother. 1987 Jan;31(1):60–66. doi: 10.1128/aac.31.1.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swedberg G. Organization of two sulfonamide resistance genes on plasmids of gram-negative bacteria. Antimicrob Agents Chemother. 1987 Feb;31(2):306–311. doi: 10.1128/aac.31.2.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swift G., McCarthy B. J., Heffron F. DNA sequence of a plasmid-encoded dihydrofolate reductase. Mol Gen Genet. 1981;181(4):441–447. doi: 10.1007/BF00428733. [DOI] [PubMed] [Google Scholar]
- Tenover F. C., Filpula D., Phillips K. L., Plorde J. J. Cloning and sequencing of a gene encoding an aminoglycoside 6'-N-acetyltransferase from an R factor of Citrobacter diversus. J Bacteriol. 1988 Jan;170(1):471–473. doi: 10.1128/jb.170.1.471-473.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vézina G., Levesque R. C. Molecular characterization of the class II multiresistance transposable element Tn1403 from Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1991 Feb;35(2):313–321. doi: 10.1128/aac.35.2.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward J. M., Grinsted J. Physical and genetic analysis of the Inc-W group plasmids R388, Sa, and R7K. Plasmid. 1982 May;7(3):239–250. doi: 10.1016/0147-619x(82)90005-1. [DOI] [PubMed] [Google Scholar]
- Wohlleben W., Arnold W., Bissonnette L., Pelletier A., Tanguay A., Roy P. H., Gamboa G. C., Barry G. F., Aubert E., Davies J. On the evolution of Tn21-like multiresistance transposons: sequence analysis of the gene (aacC1) for gentamicin acetyltransferase-3-I(AAC(3)-I), another member of the Tn21-based expression cassette. Mol Gen Genet. 1989 Jun;217(2-3):202–208. doi: 10.1007/BF02464882. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- de la Cruz F., Grinsted J. Genetic and molecular characterization of Tn21, a multiple resistance transposon from R100.1. J Bacteriol. 1982 Jul;151(1):222–228. doi: 10.1128/jb.151.1.222-228.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]



