Abstract
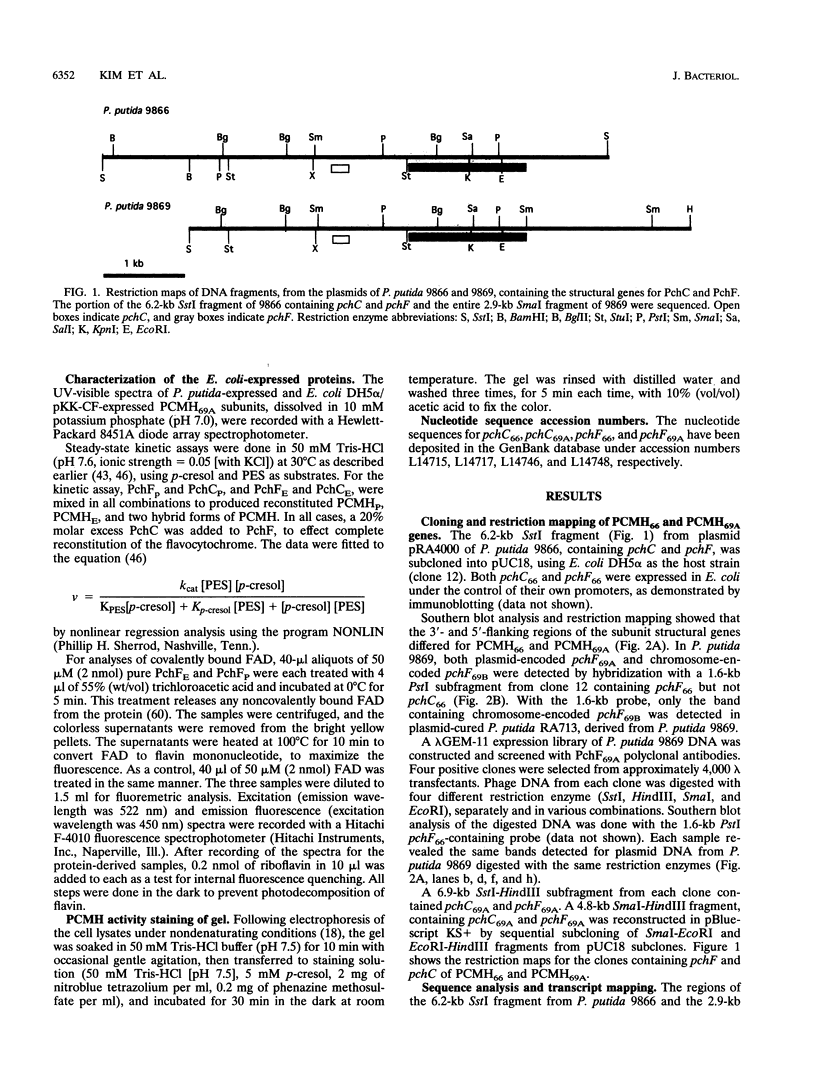
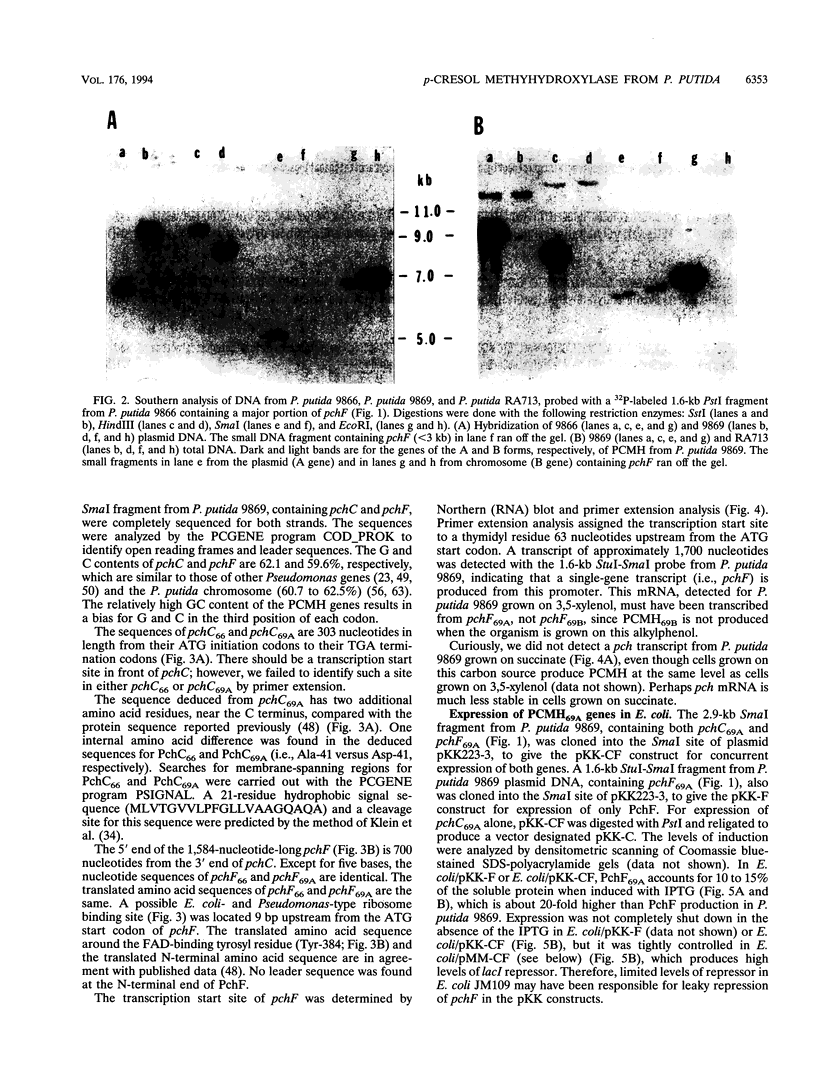
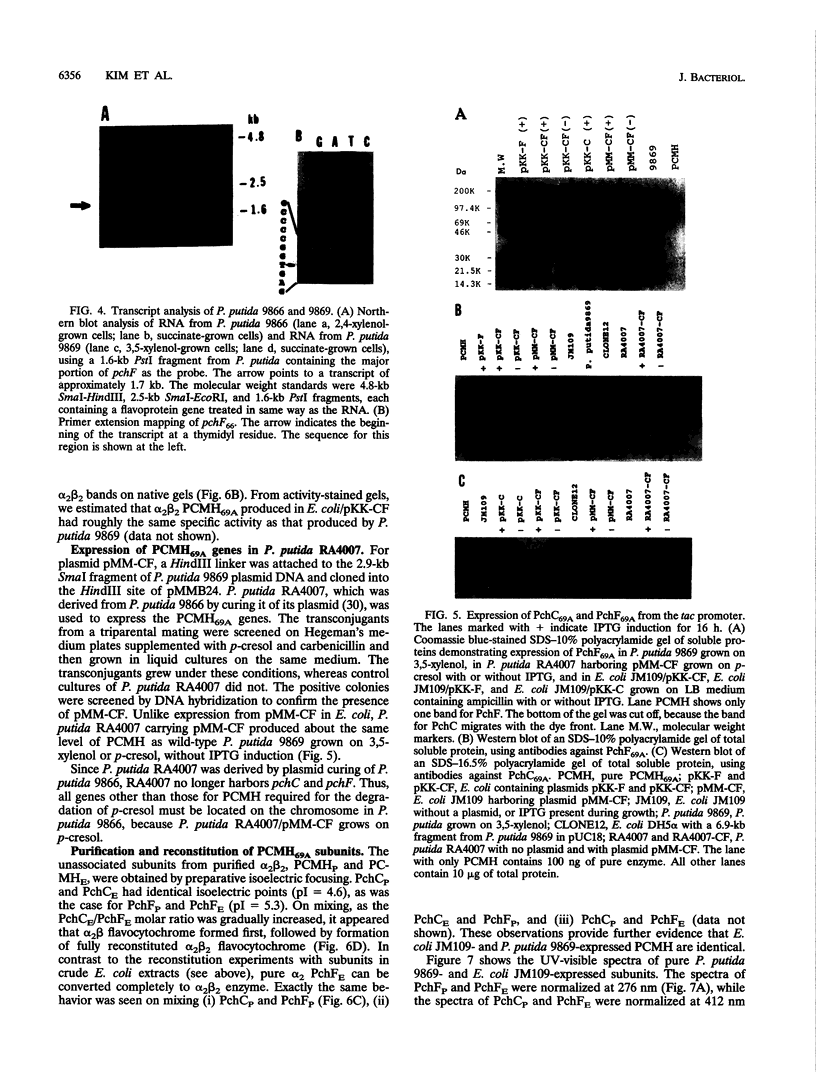
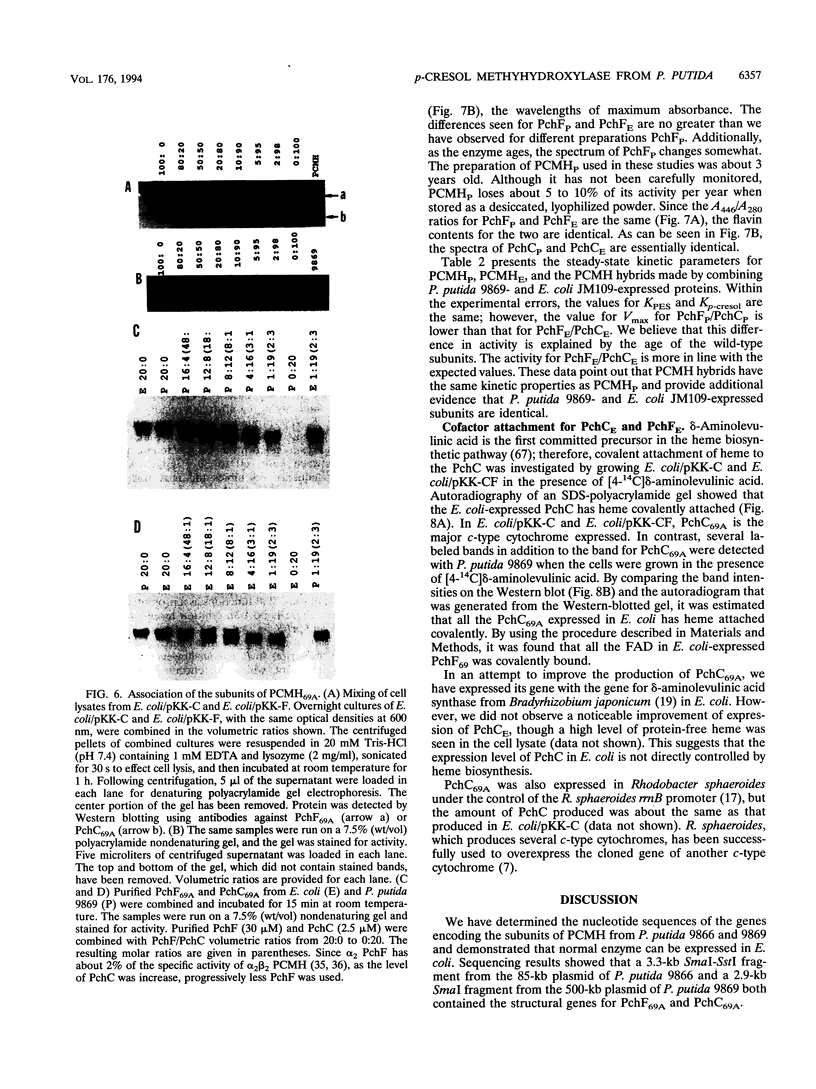
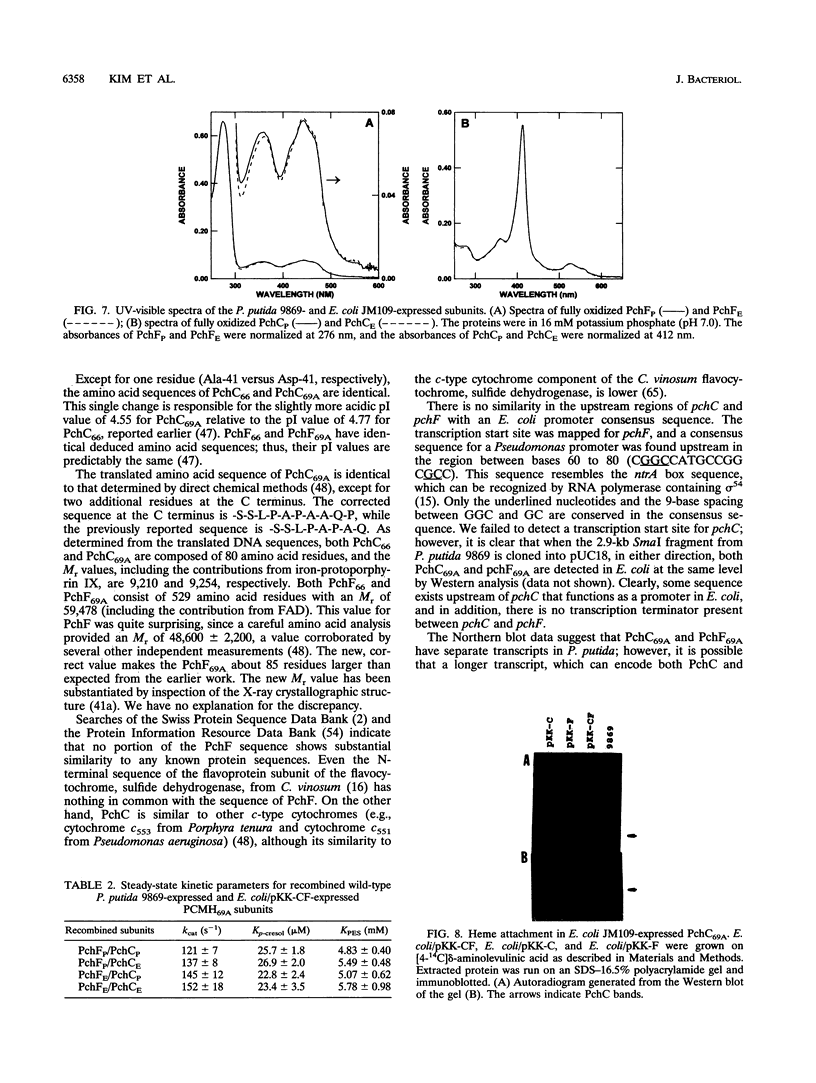
The structural genes for the flavoprotein subunit and cytochrome c subunit of p-cresol (4-methylphenol) methylhydroxylase (PCMH) from Pseudomonas putida NCIMB 9869 (National Collection of Industrial and Marine Bacteria, Aberdeen, Scotland) and P. putida NCIMB 9866 were cloned and sequenced. The genes from P.putida NCIMB 9869 were for the plasmid-encoded A form of PCMH, and the genes from P.putida NCIMB 9866 were also plasmid encoded. The nucleotide sequences of the two flavoprotein genes from P.putida NCIMB 9869 and P.putida NCIMB 9866 (pchF69A and pchF66, respectively) were the same except for 5 bases out of 1,584, and the translated amino acid sequences were identical. The nucleotide sequences of the genes for the cytochrome subunits of PCMH from the two bacteria (pchC69A and pchC66) varied by a single nucleotide in their 303-base sequences, and the translated amino acid sequences differed by a single residue at position 41 (Asp in PchC69A and Ala in PchC66). Both cytochromes had 21-residue signal sequences, as expected for periplasmic proteins, and these sequences were identical. On the other hand, no signal sequences were found for the flavoproteins.pchF69A and pchC69A were expressed, separately or together, in Escherichia coli JM109 and P.putida RA4007, with active PCMH produced in both bacteria. The E. coli-expressed flavocytochrome was purified. Our studies indicated that the E.coli-expressed subunits were identical to the subunits expressed in P.putida NCIMB 9869: molecular weights, isoelectric points, UV-visible spectra, and steady-state kinetic parameters were the same for the two sets of proteins. The subunits readily associated upon mixing two crude extracts of E.coli, one extract containing PchC69A and the other containing PchF69A. The courses of association of PchC69A and PchF69A were essentially identical for pure E. coli-expressed subunits and pure P. putida 9869-expressed subunits. E. coli-expressed PchC69A and PchF69A contained covalently bound heme and covalently bound flavin adenine dinucleotide, respectively, as the proteins expressed in nature.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bagdasarian M. M., Amann E., Lurz R., Rückert B., Bagdasarian M. Activity of the hybrid trp-lac (tac) promoter of Escherichia coli in Pseudomonas putida. Construction of broad-host-range, controlled-expression vectors. Gene. 1983 Dec;26(2-3):273–282. doi: 10.1016/0378-1119(83)90197-x. [DOI] [PubMed] [Google Scholar]
- Beckman D. L., Kranz R. G. Cytochromes c biogenesis in a photosynthetic bacterium requires a periplasmic thioredoxin-like protein. Proc Natl Acad Sci U S A. 1993 Mar 15;90(6):2179–2183. doi: 10.1073/pnas.90.6.2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaut M., Whittaker K., Valdovinos A., Ackrell B. A., Gunsalus R. P., Cecchini G. Fumarate reductase mutants of Escherichia coli that lack covalently bound flavin. J Biol Chem. 1989 Aug 15;264(23):13599–13604. [PubMed] [Google Scholar]
- Brandner J. P., McEwan A. G., Kaplan S., Donohue T. J. Expression of the Rhodobacter sphaeroides cytochrome c2 structural gene. J Bacteriol. 1989 Jan;171(1):360–368. doi: 10.1128/jb.171.1.360-368.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandsch R., Bichler V., Krauss B. Binding of FAD to 6-hydroxy-D-nicotine oxidase apoenzyme prevents degradation of the holoenzyme. Biochem J. 1989 Feb 15;258(1):187–192. doi: 10.1042/bj2580187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- COHEN-BAZIRE G., SISTROM W. R., STANIER R. Y. Kinetic studies of pigment synthesis by non-sulfur purple bacteria. J Cell Physiol. 1957 Feb;49(1):25–68. doi: 10.1002/jcp.1030490104. [DOI] [PubMed] [Google Scholar]
- Chlumsky L. J., Zhang L., Ramsey A. J., Jorns M. S. Preparation and properties of recombinant corynebacterial sarcosine oxidase: evidence for posttranslational modification during turnover with sarcosine. Biochemistry. 1993 Oct 19;32(41):11132–11142. doi: 10.1021/bi00092a024. [DOI] [PubMed] [Google Scholar]
- Decker K., Brandsch R. Flavoproteins with a covalent histidyl(N3)-8 alpha-riboflavin linkage. Biofactors. 1991 Jun;3(2):69–81. [PubMed] [Google Scholar]
- Dolata M. M., Van Beeumen J. J., Ambler R. P., Meyer T. E., Cusanovich M. A. Nucleotide sequence of the heme subunit of flavocytochrome c from the purple phototrophic bacterium, Chromatium vinosum. A 2.6-kilobase pair DNA fragment contains two multiheme cytochromes, a flavoprotein, and a homolog of human ankyrin. J Biol Chem. 1993 Jul 5;268(19):14426–14431. [PubMed] [Google Scholar]
- Dryden S. C., Kaplan S. Localization and structural analysis of the ribosomal RNA operons of Rhodobacter sphaeroides. Nucleic Acids Res. 1990 Dec 25;18(24):7267–7277. doi: 10.1093/nar/18.24.7267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerinot M. L., Chelm B. K. Bacterial delta-aminolevulinic acid synthase activity is not essential for leghemoglobin formation in the soybean/Bradyrhizobium japonicum symbiosis. Proc Natl Acad Sci U S A. 1986 Mar;83(6):1837–1841. doi: 10.1073/pnas.83.6.1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegeman G. D. Synthesis of the enzymes of the mandelate pathway by Pseudomonas putida. I. Synthesis of enzymes by the wild type. J Bacteriol. 1966 Mar;91(3):1140–1154. doi: 10.1128/jb.91.3.1140-1154.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S. Unidirectional digestion with exonuclease III in DNA sequence analysis. Methods Enzymol. 1987;155:156–165. doi: 10.1016/0076-6879(87)55014-5. [DOI] [PubMed] [Google Scholar]
- Holloway B. W., Morgan A. F. Genome organization in Pseudomonas. Annu Rev Microbiol. 1986;40:79–105. doi: 10.1146/annurev.mi.40.100186.000455. [DOI] [PubMed] [Google Scholar]
- Hopper D. J., Bossert I. D., Rhodes-Roberts M. E. p-cresol methylhydroxylase from a denitrifying bacterium involved in anaerobic degradation of p-cresol. J Bacteriol. 1991 Feb;173(3):1298–1301. doi: 10.1128/jb.173.3.1298-1301.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopper D. J., Jones M. R., Causer M. J. Periplasmic location of p-cresol methylhydroxylase in Pseudomonas putida. FEBS Lett. 1985 Mar 25;182(2):485–488. doi: 10.1016/0014-5793(85)80359-8. [DOI] [PubMed] [Google Scholar]
- Hopper D. J., Kemp P. D. Regulation of enzymes of the 3,5-xylenol-degradative pathway in Pseudomonas putida: evidence for a plasmid. J Bacteriol. 1980 Apr;142(1):21–26. doi: 10.1128/jb.142.1.21-26.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopper D. J. Redox potential of the cytochrome c in the flavocytochrome p-cresol methylhydroxylase. FEBS Lett. 1983 Sep 5;161(1):100–102. doi: 10.1016/0014-5793(83)80738-8. [DOI] [PubMed] [Google Scholar]
- Hopper D. J., Taylor D. G. The purification and properties of p-cresol-(acceptor) oxidoreductase (hydroxylating), a flavocytochrome from Pseudomonas putida. Biochem J. 1977 Oct 1;167(1):155–162. doi: 10.1042/bj1670155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones K. A., Yamamoto K. R., Tjian R. Two distinct transcription factors bind to the HSV thymidine kinase promoter in vitro. Cell. 1985 Sep;42(2):559–572. doi: 10.1016/0092-8674(85)90113-8. [DOI] [PubMed] [Google Scholar]
- Keat M. J., Hopper D. J. P-cresol and 3,5-xylenol methylhydroxylases in Pseudomonas putida N.C.I.B. 9896. Biochem J. 1978 Nov 1;175(2):649–658. doi: 10.1042/bj1750649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein P., Kanehisa M., DeLisi C. The detection and classification of membrane-spanning proteins. Biochim Biophys Acta. 1985 May 28;815(3):468–476. doi: 10.1016/0005-2736(85)90375-x. [DOI] [PubMed] [Google Scholar]
- Koerber S. C., Hopper D. J., McIntire W. S., Singer T. P. Formation and properties of flavoprotein-cytochrome hybrids by recombination of subunits from different species. Biochem J. 1985 Oct 15;231(2):383–387. doi: 10.1042/bj2310383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koerber S. C., McIntire W., Bohmont C., Singer T. P. Resolution of the flavocytochrome p-cresol methylhydroxylase into subunits and reconstitution of the enzyme. Biochemistry. 1985 Sep 10;24(19):5276–5280. doi: 10.1021/bi00340a048. [DOI] [PubMed] [Google Scholar]
- Koronakis V., Koronakis E., Hughes C. Isolation and analysis of the C-terminal signal directing export of Escherichia coli hemolysin protein across both bacterial membranes. EMBO J. 1989 Feb;8(2):595–605. doi: 10.1002/j.1460-2075.1989.tb03414.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuchler K., Thorner J. Secretion of peptides and proteins lacking hydrophobic signal sequences: the role of adenosine triphosphate-driven membrane translocators. Endocr Rev. 1992 Aug;13(3):499–514. doi: 10.1210/edrv-13-3-499. [DOI] [PubMed] [Google Scholar]
- Ladin B. F., Accuroso E. M., Mielenz J. R., Wilson C. R. A rapid procedure for isolating RNA from small-scale Bacillus cultures. Biotechniques. 1992 May;12(5):672–676. [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lovley D. R., Lonergan D. J. Anaerobic Oxidation of Toluene, Phenol, and p-Cresol by the Dissimilatory Iron-Reducing Organism, GS-15. Appl Environ Microbiol. 1990 Jun;56(6):1858–1864. doi: 10.1128/aem.56.6.1858-1864.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews F. S., Chen Z. W., Bellamy H. D., McIntire W. S. Three-dimensional structure of p-cresol methylhydroxylase (flavocytochrome c) from Pseudomonas putida at 3.0-A resolution. Biochemistry. 1991 Jan 8;30(1):238–247. doi: 10.1021/bi00215a034. [DOI] [PubMed] [Google Scholar]
- McIntire W. S., Hopper D. J., Singer T. P. Steady-state and stopped-flow kinetic measurements of the primary deuterium isotope effect in the reaction catalyzed by p-cresol methylhydroxylase. Biochemistry. 1987 Jun 30;26(13):4107–4117. doi: 10.1021/bi00387a055. [DOI] [PubMed] [Google Scholar]
- McIntire W. S. Trimethylamine dehydrogenase from bacterium W3A1. Methods Enzymol. 1990;188:250–260. doi: 10.1016/0076-6879(90)88042-9. [DOI] [PubMed] [Google Scholar]
- McIntire W., Edmondson D. E., Hopper D. J., Singer T. P. 8 alpha-(O-Tyrosyl)flavin adenine dinucleotide, the prosthetic group of bacterial p-cresol methylhydroxylase. Biochemistry. 1981 May 26;20(11):3068–3075. doi: 10.1021/bi00514a013. [DOI] [PubMed] [Google Scholar]
- McIntire W., Hopper D. J., Craig J. C., Everhart E. T., Webster R. V., Causer M. J., Singer T. P. Stereochemistry of 1-(4'-hydroxyphenyl)ethanol produced by hydroxylation of 4-ethylphenol by p-cresol methylhydroxylase. Biochem J. 1984 Dec 1;224(2):617–621. doi: 10.1042/bj2240617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntire W., Singer T. P., Smith A. J., Mathews F. S. Amino acid and sequence analysis of the cytochrome and flavoprotein subunits of p-cresol methylhydroxylase. Biochemistry. 1986 Oct 7;25(20):5975–5981. doi: 10.1021/bi00368a021. [DOI] [PubMed] [Google Scholar]
- Misra T. K., Brown N. L., Fritzinger D. C., Pridmore R. D., Barnes W. M., Haberstroh L., Silver S. Mercuric ion-resistance operons of plasmid R100 and transposon Tn501: the beginning of the operon including the regulatory region and the first two structural genes. Proc Natl Acad Sci U S A. 1984 Oct;81(19):5975–5979. doi: 10.1073/pnas.81.19.5975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai C., Kagamiyama H., Nozaki M., Nakazawa T., Inouye S., Ebina Y., Nakazawa A. Complete nucleotide sequence of the metapyrocatechase gene on the TOI plasmid of Pseudomonas putida mt-2. J Biol Chem. 1983 Mar 10;258(5):2923–2928. [PubMed] [Google Scholar]
- O'Reilly K. T., Crawford R. L. Kinetics of p-cresol degradation by an immobilized Pseudomonas sp. Appl Environ Microbiol. 1989 Apr;55(4):866–870. doi: 10.1128/aem.55.4.866-870.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page M. D., Ferguson S. J. Apo forms of cytochrome c550 and cytochrome cd1 are translocated to the periplasm of Paracoccus denitrificans in the absence of haem incorporation caused either mutation or inhibition of haem synthesis. Mol Microbiol. 1990 Jul;4(7):1181–1192. doi: 10.1111/j.1365-2958.1990.tb00693.x. [DOI] [PubMed] [Google Scholar]
- Pemberton J. M., Clark A. J. Detection and characterization of plasmids in Pseudomonas aeruginosa strain PAO. J Bacteriol. 1973 Apr;114(1):424–433. doi: 10.1128/jb.114.1.424-433.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeve C. D., Carver M. A., Hopper D. J. The purification and characterization of 4-ethylphenol methylenehydroxylase, a flavocytochrome from Pseudomonas putida JD1. Biochem J. 1989 Oct 15;263(2):431–437. doi: 10.1042/bj2630431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothmel R. K., Chakrabarty A. M., Berry A., Darzins A. Genetic systems in Pseudomonas. Methods Enzymol. 1991;204:485–514. doi: 10.1016/0076-6879(91)04025-j. [DOI] [PubMed] [Google Scholar]
- Rudolphi A., Tschech A., Fuchs G. Anaerobic degradation of cresols by denitrifying bacteria. Arch Microbiol. 1991;155(3):238–248. doi: 10.1007/BF00252207. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel L. M. Quantitative determination of noncovalently bound flavins: types and methods of analysis. Methods Enzymol. 1978;53:419–429. doi: 10.1016/s0076-6879(78)53046-2. [DOI] [PubMed] [Google Scholar]
- Singer T. P., McIntire W. S. Covalent attachment of flavin to flavoproteins: occurrence, assay, and synthesis. Methods Enzymol. 1984;106:369–378. doi: 10.1016/0076-6879(84)06039-0. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Stanier R. Y., Palleroni N. J., Doudoroff M. The aerobic pseudomonads: a taxonomic study. J Gen Microbiol. 1966 May;43(2):159–271. doi: 10.1099/00221287-43-2-159. [DOI] [PubMed] [Google Scholar]
- Towbin H., Staehelin T., Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Beeumen J. J., Demol H., Samyn B., Bartsch R. G., Meyer T. E., Dolata M. M., Cusanovich M. A. Covalent structure of the diheme cytochrome subunit and amino-terminal sequence of the flavoprotein subunit of flavocytochrome c from Chromatium vinosum. J Biol Chem. 1991 Jul 15;266(20):12921–12931. [PubMed] [Google Scholar]
- Wickner W., Driessen A. J., Hartl F. U. The enzymology of protein translocation across the Escherichia coli plasma membrane. Annu Rev Biochem. 1991;60:101–124. doi: 10.1146/annurev.bi.60.070191.000533. [DOI] [PubMed] [Google Scholar]
- Wyckoff M., Rodbard D., Chrambach A. Polyacrylamide gel electrophoresis in sodium dodecyl sulfate-containing buffers using multiphasic buffer systems: properties of the stack, valid Rf- measurement, and optimized procedure. Anal Biochem. 1977 Apr;78(2):459–482. doi: 10.1016/0003-2697(77)90107-5. [DOI] [PubMed] [Google Scholar]
- von Wachenfeldt C., Hederstedt L. Bacillus subtilis 13-kilodalton cytochrome c-550 encoded by cccA consists of a membrane-anchor and a heme domain. J Biol Chem. 1990 Aug 15;265(23):13939–13948. [PubMed] [Google Scholar]
- von Wachenfeldt C., Hederstedt L. Bacillus subtilis holo-cytochrome c-550 can be synthesised in aerobic Escherichia coli. FEBS Lett. 1990 Sep 17;270(1-2):147–151. doi: 10.1016/0014-5793(90)81255-m. [DOI] [PubMed] [Google Scholar]