Abstract
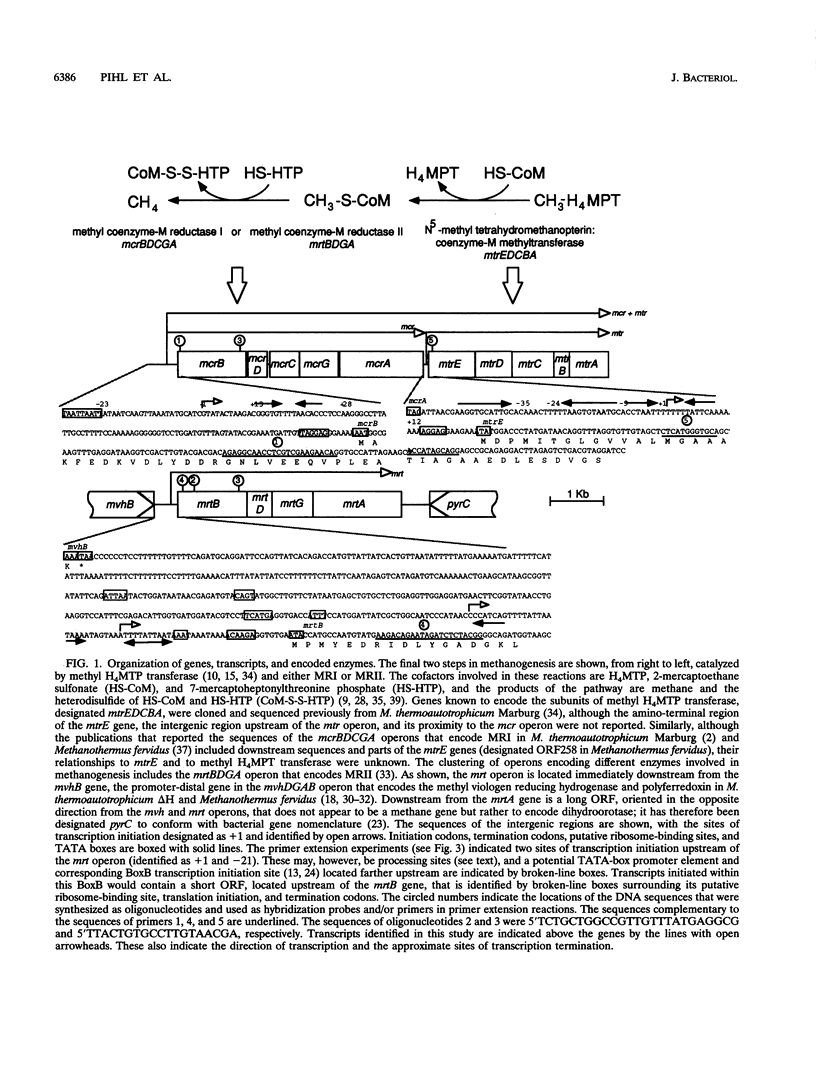
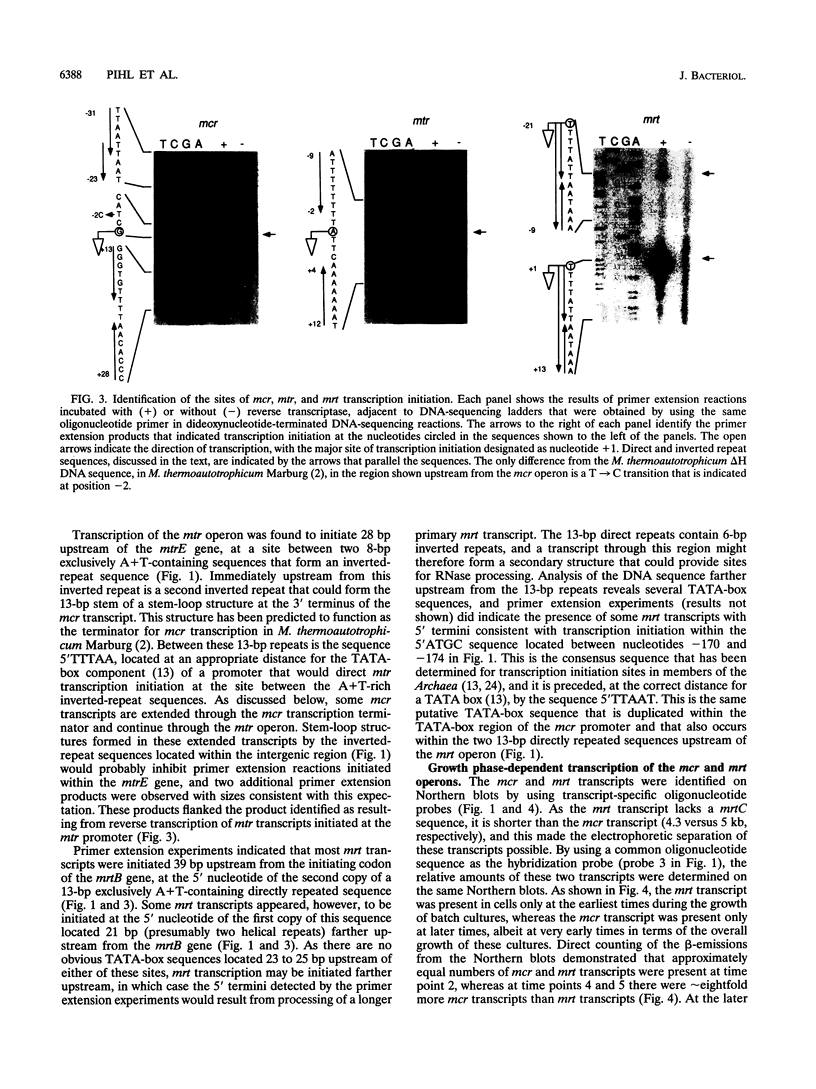
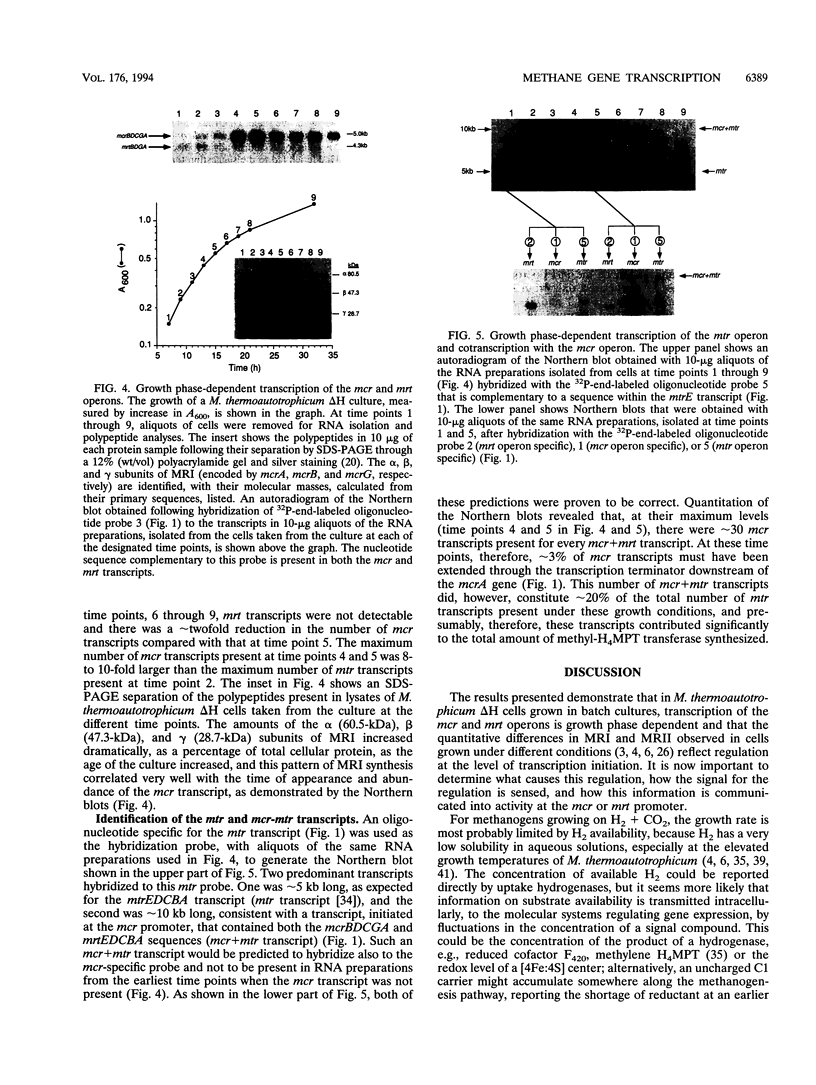
The genes encoding the two isoenzymes of methyl coenzyme M reductase (MRI and MRII) in Methanobacterium thermoautotrophicum delta H have been cloned and sequenced. The MRI-encoding mcr operon (mcrBDCGA) has been located immediately upstream from the mtr operon (mtrEDCBA) that encodes N5-methyltetrahydromethanopterin:coenzyme M methyltransferase, the enzyme that catalyzes the step preceding the MR-catalyzed reaction in methanogenesis. The MRII-encoding mrt operon (mrtBDGA) has been located between the operon that encodes the methyl viologen-reducing hydrogenase and an open reading frame (designated pyrC) predicted to encode dihydroorotase. Surprisingly, the mrt operon has been found to contain only four genes (mrtBDGA), lacking the equivalent of the mcrC gene that is present in all mcr operons. A protocol that isolates transcripts intact from M. thermoautotrophicum delta H cells has been developed and used, with primer extension and Northern (RNA) blot procedures, to identify the sites of transcription initiation upstream of the mcr, mrt, and mtr operons and to determine the relative numbers of these transcripts in cells at different growth stages. Transcription of the mrt operon was found to occur only at early times in batch cultures and was then replaced by transcription of the mcr operon. Transcripts of the mtr operon were detectable at all times; however, at early times, all mtr transcripts were initiated at the mtr promoter, whereas at later times, during mcr transcription, approximately 3% of mcr transcripts were extended to generate mcr plus mtr transcripts that constituted approximately 20% of all mtr transcripts present.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bertram P. A., Schmitz R. A., Linder D., Thauer R. K. Tungstate can substitute for molybdate in sustaining growth of Methanobacterium thermoautotrophicum. Identification and characterization of a tungsten isoenzyme of formylmethanofuran dehydrogenase. Arch Microbiol. 1994;161(3):220–228. doi: 10.1007/BF00248696. [DOI] [PubMed] [Google Scholar]
- Bokranz M., Bäumner G., Allmansberger R., Ankel-Fuchs D., Klein A. Cloning and characterization of the methyl coenzyme M reductase genes from Methanobacterium thermoautotrophicum. J Bacteriol. 1988 Feb;170(2):568–577. doi: 10.1128/jb.170.2.568-577.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonacker L. G., Baudner S., Mörschel E., Böcher R., Thauer R. K. Properties of the two isoenzymes of methyl-coenzyme M reductase in Methanobacterium thermoautotrophicum. Eur J Biochem. 1993 Oct 15;217(2):587–595. doi: 10.1111/j.1432-1033.1993.tb18281.x. [DOI] [PubMed] [Google Scholar]
- Bonacker L. G., Baudner S., Thauer R. K. Differential expression of the two methyl-coenzyme M reductases in Methanobacterium thermoautotrophicum as determined immunochemically via isoenzyme-specific antisera. Eur J Biochem. 1992 May 15;206(1):87–92. doi: 10.1111/j.1432-1033.1992.tb16904.x. [DOI] [PubMed] [Google Scholar]
- Brawerman G. mRNA decay: finding the right targets. Cell. 1989 Apr 7;57(1):9–10. doi: 10.1016/0092-8674(89)90166-9. [DOI] [PubMed] [Google Scholar]
- Brenner M. C., Ma L., Johnson M. K., Scott R. A. Spectroscopic characterization of the alternate form of S-methylcoenzyme M reductase from Methanobacterium thermoautotrophicum (strain delta H). Biochim Biophys Acta. 1992 Apr 8;1120(2):160–166. doi: 10.1016/0167-4838(92)90264-e. [DOI] [PubMed] [Google Scholar]
- Ellermann J., Hedderich R., Böcher R., Thauer R. K. The final step in methane formation. Investigations with highly purified methyl-CoM reductase (component C) from Methanobacterium thermoautotrophicum (strain Marburg). Eur J Biochem. 1988 Mar 15;172(3):669–677. doi: 10.1111/j.1432-1033.1988.tb13941.x. [DOI] [PubMed] [Google Scholar]
- Ellermann J., Rospert S., Thauer R. K., Bokranz M., Klein A., Voges M., Berkessel A. Methyl-coenzyme-M reductase from Methanobacterium thermoautotrophicum (strain Marburg). Purity, activity and novel inhibitors. Eur J Biochem. 1989 Sep 1;184(1):63–68. doi: 10.1111/j.1432-1033.1989.tb14990.x. [DOI] [PubMed] [Google Scholar]
- Ferry J. G. Biochemistry of methanogenesis. Crit Rev Biochem Mol Biol. 1992;27(6):473–503. doi: 10.3109/10409239209082570. [DOI] [PubMed] [Google Scholar]
- Gärtner P., Ecker A., Fischer R., Linder D., Fuchs G., Thauer R. K. Purification and properties of N5-methyltetrahydromethanopterin:coenzyme M methyltransferase from Methanobacterium thermoautotrophicum. Eur J Biochem. 1993 Apr 1;213(1):537–545. doi: 10.1111/j.1432-1033.1993.tb17792.x. [DOI] [PubMed] [Google Scholar]
- Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983 Jun 5;166(4):557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- Hausner W., Frey G., Thomm M. Control regions of an archaeal gene. A TATA box and an initiator element promote cell-free transcription of the tRNA(Val) gene of Methanococcus vannielii. J Mol Biol. 1991 Dec 5;222(3):495–508. doi: 10.1016/0022-2836(91)90492-o. [DOI] [PubMed] [Google Scholar]
- Hennigan A. N., Reeve J. N. mRNAs in the methanogenic archaeon Methanococcus vannielii: numbers, half-lives and processing. Mol Microbiol. 1994 Feb;11(4):655–670. doi: 10.1111/j.1365-2958.1994.tb00344.x. [DOI] [PubMed] [Google Scholar]
- Kengen S. W., Daas P. J., Duits E. F., Keltjens J. T., van der Drift C., Vogels G. D. Isolation of a 5-hydroxybenzimidazolyl cobamide-containing enzyme involved in the methyltetrahydromethanopterin: coenzyme M methyltransferase reaction in Methanobacterium thermoautotrophicum. Biochim Biophys Acta. 1992 Feb 1;1118(3):249–260. doi: 10.1016/0167-4838(92)90282-i. [DOI] [PubMed] [Google Scholar]
- Kengen S. W., von den Hoff H. W., Keltjens J. T., van der Drift C., Vogels G. D. Hydrolysis and reduction of factor 390 by cell extracts of Methanobacterium thermoautotrophicum (strain delta H). J Bacteriol. 1991 Apr;173(7):2283–2288. doi: 10.1128/jb.173.7.2283-2288.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein A., Allmansberger R., Bokranz M., Knaub S., Müller B., Muth E. Comparative analysis of genes encoding methyl coenzyme M reductase in methanogenic bacteria. Mol Gen Genet. 1988 Aug;213(2-3):409–420. doi: 10.1007/BF00339610. [DOI] [PubMed] [Google Scholar]
- Lehmacher A., Klenk H. P. Characterization and phylogeny of mcrII, a gene cluster encoding an isoenzyme of methyl coenzyme M reductase from hyperthermophilic Methanothermus fervidus. Mol Gen Genet. 1994 Apr;243(2):198–206. doi: 10.1007/BF00280317. [DOI] [PubMed] [Google Scholar]
- Montzka K. A., Steitz J. A. Additional low-abundance human small nuclear ribonucleoproteins: U11, U12, etc. Proc Natl Acad Sci U S A. 1988 Dec;85(23):8885–8889. doi: 10.1073/pnas.85.23.8885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka A., Sugimoto K., Sasaki H., Takanami M. An in vitro method generating base substitutions in preselected regions of plasmid DNA: application to structural analysis of the replication origin of the Escherichia coli K-12 chromosome. Gene. 1982 Jul-Aug;19(1):59–69. doi: 10.1016/0378-1119(82)90189-5. [DOI] [PubMed] [Google Scholar]
- Petersen C. Control of functional mRNA stability in bacteria: multiple mechanisms of nucleolytic and non-nucleolytic inactivation. Mol Microbiol. 1992 Feb;6(3):277–282. doi: 10.1111/j.1365-2958.1992.tb01469.x. [DOI] [PubMed] [Google Scholar]
- Polacheck I., Cabib E. A simple procedure for protein determination by the Lowry method in dilute solutions and in the presence of interfering substances. Anal Biochem. 1981 Nov 1;117(2):311–314. doi: 10.1016/0003-2697(81)90784-3. [DOI] [PubMed] [Google Scholar]
- Quinn C. L., Stephenson B. T., Switzer R. L. Functional organization and nucleotide sequence of the Bacillus subtilis pyrimidine biosynthetic operon. J Biol Chem. 1991 May 15;266(14):9113–9127. [PubMed] [Google Scholar]
- Reeve J. N., Beckler G. S., Cram D. S., Hamilton P. T., Brown J. W., Krzycki J. A., Kolodziej A. F., Alex L., Orme-Johnson W. H., Walsh C. T. A hydrogenase-linked gene in Methanobacterium thermoautotrophicum strain delta H encodes a polyferredoxin. Proc Natl Acad Sci U S A. 1989 May;86(9):3031–3035. doi: 10.1073/pnas.86.9.3031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeve J. N. Molecular biology of methanogens. Annu Rev Microbiol. 1992;46:165–191. doi: 10.1146/annurev.mi.46.100192.001121. [DOI] [PubMed] [Google Scholar]
- Rospert S., Linder D., Ellermann J., Thauer R. K. Two genetically distinct methyl-coenzyme M reductases in Methanobacterium thermoautotrophicum strain Marburg and delta H. Eur J Biochem. 1990 Dec 27;194(3):871–877. doi: 10.1111/j.1432-1033.1990.tb19481.x. [DOI] [PubMed] [Google Scholar]
- Rouvière P. E., Bobik T. A., Wolfe R. S. Reductive activation of the methyl coenzyme M methylreductase system of Methanobacterium thermoautotrophicum delta H. J Bacteriol. 1988 Sep;170(9):3946–3952. doi: 10.1128/jb.170.9.3946-3952.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouvière P. E., Wolfe R. S. Novel biochemistry of methanogenesis. J Biol Chem. 1988 Jun 15;263(17):7913–7916. [PubMed] [Google Scholar]
- Steigerwald V. J., Beckler G. S., Reeve J. N. Conservation of hydrogenase and polyferredoxin structures in the hyperthermophilic archaebacterium Methanothermus fervidus. J Bacteriol. 1990 Aug;172(8):4715–4718. doi: 10.1128/jb.172.8.4715-4718.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stettler R., Leisinger T. Physical map of the Methanobacterium thermoautotrophicum Marburg chromosome. J Bacteriol. 1992 Nov;174(22):7227–7234. doi: 10.1128/jb.174.22.7227-7234.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stupperich E., Juza A., Hoppert M., Mayer F. Cloning, sequencing and immunological characterization of the corrinoid-containing subunit of the N5-methyltetrahydromethanopterin: coenzyme-M methyltransferase from Methanobacterium thermoautotrophicum. Eur J Biochem. 1993 Oct 1;217(1):115–121. doi: 10.1111/j.1432-1033.1993.tb18225.x. [DOI] [PubMed] [Google Scholar]
- Weil C. F., Cram D. S., Sherf B. A., Reeve J. N. Structure and comparative analysis of the genes encoding component C of methyl coenzyme M reductase in the extremely thermophilic archaebacterium Methanothermus fervidus. J Bacteriol. 1988 Oct;170(10):4718–4726. doi: 10.1128/jb.170.10.4718-4726.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weil C. F., Sherf B. A., Reeve J. N. A comparison of the methyl reductase genes and gene products. Can J Microbiol. 1989 Jan;35(1):101–108. doi: 10.1139/m89-016. [DOI] [PubMed] [Google Scholar]
- Weiss D. S., Thauer R. K. Methanogenesis and the unity of biochemistry. Cell. 1993 Mar 26;72(6):819–822. doi: 10.1016/0092-8674(93)90570-g. [DOI] [PubMed] [Google Scholar]
- Woo G. J., Wasserfallen A., Wolfe R. S. Methyl viologen hydrogenase II, a new member of the hydrogenase family from Methanobacterium thermoautotrophicum delta H. J Bacteriol. 1993 Sep;175(18):5970–5977. doi: 10.1128/jb.175.18.5970-5977.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]