Abstract
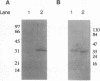
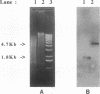
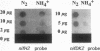
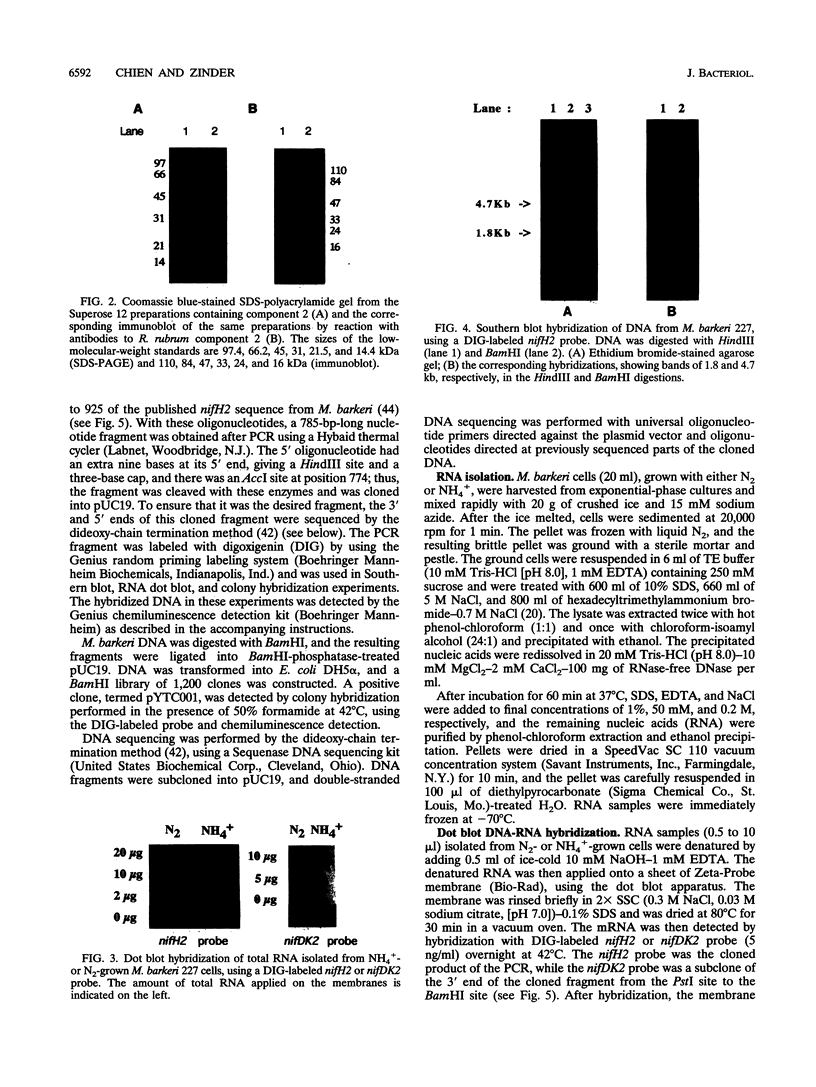
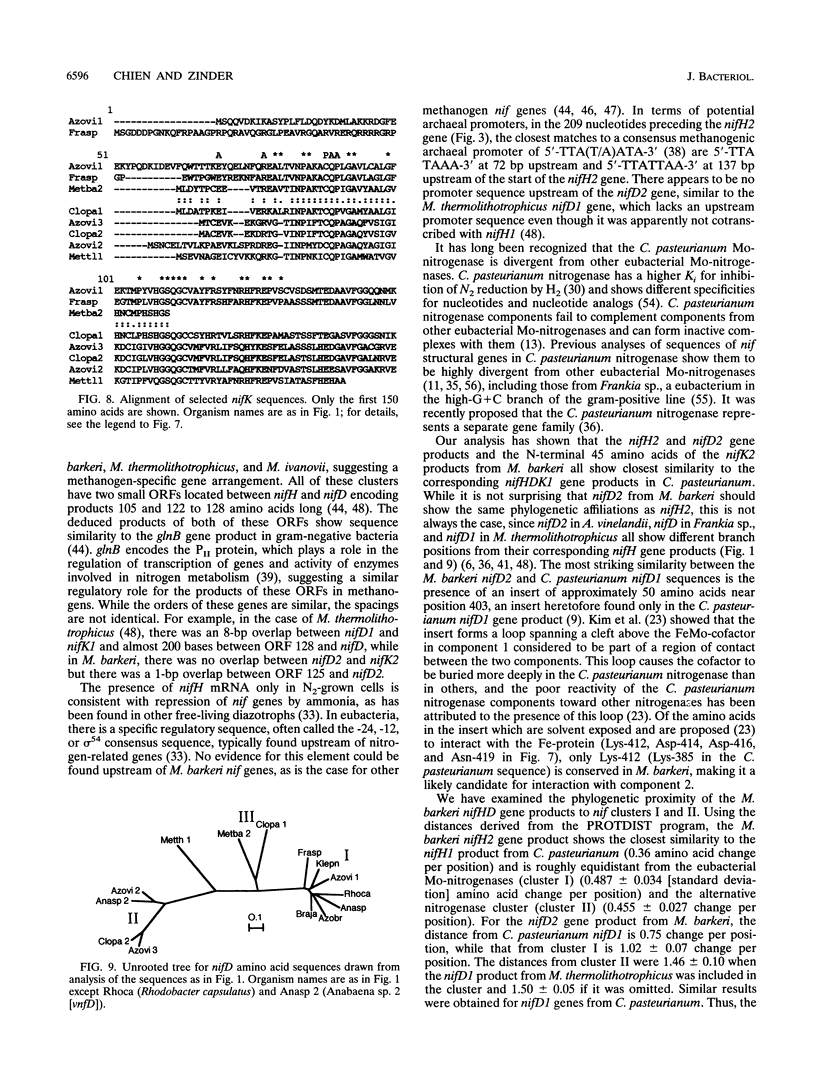
L. Sibold, M. Henriquet, O. Possot, and J.-P. Aubert (Res. Microbiol. 142:5-12, 1991) cloned and sequenced two nifH-homologous open reading frames (ORFs) from Methanosarcina barkeri 227. Phylogenetic analysis of the deduced amino acid sequences of the nifH ORFs from M. barkeri showed that nifH1 clusters with nifH genes from alternative nitrogenases, while nifH2 clusters with nifH1 from the gram-positive eubacterium Clostridium pasteurianum. The N-terminal sequence of the purified nitrogenase component 2 (the nifH gene product) from M. barkeri was identical with that predicted for nifH2, and dot blot analysis of RNA transcripts indicated that nifH2 (and nifDK2) was expressed in M. barkeri when grown diazotrophically in Mo-containing medium. To obtain nifD2 from M. barkeri, a 4.7-kbp BamHI fragment of M. barkeri DNA was cloned which contained at least five ORFs, including nifH2, ORF105, and ORF125 (previously described by Sibold et al.), as well as nifD2 and part of nifK2. ORFnifD2 is 1,596 bp long and encodes 532 amino acid residues, while the nifK2 fragment is 135 bp long. The deduced amino acid sequences for nifD2 and the nifK2 fragment from M. barkeri cluster most closely with the corresponding nifDK1 gene products from C. pasteurianum. The predicted M. barkeri nifD2 product contains a 50-amino acid insert near the C terminus which has previously been found only in the clostridial nifD1 product. Previous biochemical and sequencing evidence indicates that the C. pasteurianum nitrogenase is the most divergent of known eubacterial Mo-nitrogenases, most likely representing a distinct nif gene family, which now also contains M. barkeri as a member. The similarity between the methanogen and clostridial nif sequences is especially intriguing in light of the recent findings of sequence similarities between gene products from archaea and from low-G+C gram-positive eubacteria for glutamate dehydrogenase, glutamine synthetase I, and heat shock protein 70. It is not clear whether this similarity is due to horizontal gene transfer or to the resemblance of the M. barkeri and C. pasteurianum nitrogenase sequences to an ancestral nitrogenase.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Balch W. E., Fox G. E., Magrum L. J., Woese C. R., Wolfe R. S. Methanogens: reevaluation of a unique biological group. Microbiol Rev. 1979 Jun;43(2):260–296. doi: 10.1128/mr.43.2.260-296.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belay N., Sparling R., Choi B. S., Roberts M., Roberts J. E., Daniels L. Physiological and 15N-NMR analysis of molecular nitrogen fixation by Methanococcus thermolithotrophicus, Methanobacterium bryantii and Methanospirillum hungatei. Biochim Biophys Acta. 1988 Oct 7;971(3):233–245. doi: 10.1016/0167-4889(88)90138-3. [DOI] [PubMed] [Google Scholar]
- Belay N., Sparling R., Daniels L. Dinitrogen fixation by a thermophilic methanogenic bacterium. Nature. 1984 Nov 15;312(5991):286–288. doi: 10.1038/312286a0. [DOI] [PubMed] [Google Scholar]
- Benachenhou-Lahfa N., Forterre P., Labedan B. Evolution of glutamate dehydrogenase genes: evidence for two paralogous protein families and unusual branching patterns of the archaebacteria in the universal tree of life. J Mol Evol. 1993 Apr;36(4):335–346. doi: 10.1007/BF00182181. [DOI] [PubMed] [Google Scholar]
- Bishop P. E., Jarlenski D. M., Hetherington D. R. Evidence for an alternative nitrogen fixation system in Azotobacter vinelandii. Proc Natl Acad Sci U S A. 1980 Dec;77(12):7342–7346. doi: 10.1073/pnas.77.12.7342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke D. H., Hearst J. E., Sidow A. Early evolution of photosynthesis: clues from nitrogenase and chlorophyll iron proteins. Proc Natl Acad Sci U S A. 1993 Aug 1;90(15):7134–7138. doi: 10.1073/pnas.90.15.7134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerich D. W., Burris R. H. Complementary functioning of the component proteins of nitrogenase from several bacteria. J Bacteriol. 1978 Jun;134(3):936–943. doi: 10.1128/jb.134.3.936-943.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fallik E., Chan Y. K., Robson R. L. Detection of alternative nitrogenases in aerobic gram-negative nitrogen-fixing bacteria. J Bacteriol. 1991 Jan;173(1):365–371. doi: 10.1128/jb.173.1.365-371.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fallik E., Hartel P. G., Robson R. L. Presence of a Vanadium Nitrogenase in Azotobacter paspali. Appl Environ Microbiol. 1993 Jun;59(6):1883–1886. doi: 10.1128/aem.59.6.1883-1886.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgiadis M. M., Komiya H., Chakrabarti P., Woo D., Kornuc J. J., Rees D. C. Crystallographic structure of the nitrogenase iron protein from Azotobacter vinelandii. Science. 1992 Sep 18;257(5077):1653–1659. doi: 10.1126/science.1529353. [DOI] [PubMed] [Google Scholar]
- Gupta R. S., Golding G. B. Evolution of HSP70 gene and its implications regarding relationships between archaebacteria, eubacteria, and eukaryotes. J Mol Evol. 1993 Dec;37(6):573–582. doi: 10.1007/BF00182743. [DOI] [PubMed] [Google Scholar]
- Jarrell K. F., Faguy D., Hebert A. M., Kalmokoff M. L. A general method of isolating high molecular weight DNA from methanogenic archaea (archaebacteria). Can J Microbiol. 1992 Jan;38(1):65–68. doi: 10.1139/m92-010. [DOI] [PubMed] [Google Scholar]
- Kim J., Woo D., Rees D. C. X-ray crystal structure of the nitrogenase molybdenum-iron protein from Clostridium pasteurianum at 3.0-A resolution. Biochemistry. 1993 Jul 20;32(28):7104–7115. doi: 10.1021/bi00079a006. [DOI] [PubMed] [Google Scholar]
- Kumada Y., Benson D. R., Hillemann D., Hosted T. J., Rochefort D. A., Thompson C. J., Wohlleben W., Tateno Y. Evolution of the glutamine synthetase gene, one of the oldest existing and functioning genes. Proc Natl Acad Sci U S A. 1993 Apr 1;90(7):3009–3013. doi: 10.1073/pnas.90.7.3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehman L. J., Roberts G. P. Identification of an alternative nitrogenase system in Rhodospirillum rubrum. J Bacteriol. 1991 Sep;173(18):5705–5711. doi: 10.1128/jb.173.18.5705-5711.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobo A. L., Zinder S. H. Diazotrophy and Nitrogenase Activity in the Archaebacterium Methanosarcina barkeri 227. Appl Environ Microbiol. 1988 Jul;54(7):1656–1661. doi: 10.1128/aem.54.7.1656-1661.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobo A. L., Zinder S. H. Nitrogenase in the archaebacterium Methanosarcina barkeri 227. J Bacteriol. 1990 Dec;172(12):6789–6796. doi: 10.1128/jb.172.12.6789-6796.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockshin A., Burris R. H. Inhibitors of nitrogen fixation in extracts from Clostridium pasteurianum. Biochim Biophys Acta. 1965 Nov 15;111(1):1–10. doi: 10.1016/0304-4165(65)90466-6. [DOI] [PubMed] [Google Scholar]
- Meile L., Stettler R., Banholzer R., Kotik M., Leisinger T. Tryptophan gene cluster of Methanobacterium thermoautotrophicum Marburg: molecular cloning and nucleotide sequence of a putative trpEGCFBAD operon. J Bacteriol. 1991 Aug;173(16):5017–5023. doi: 10.1128/jb.173.16.5017-5023.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Normand P., Bousquet J. Phylogeny of nitrogenase sequences in Frankia and other nitrogen-fixing microorganisms. J Mol Evol. 1989 Nov;29(5):436–447. doi: 10.1007/BF02602914. [DOI] [PubMed] [Google Scholar]
- Normand P., Gouy M., Cournoyer B., Simonet P. Nucleotide sequence of nifD from Frankia alni strain ArI3: phylogenetic inferences. Mol Biol Evol. 1992 May;9(3):495–506. doi: 10.1093/oxfordjournals.molbev.a040737. [DOI] [PubMed] [Google Scholar]
- Reeve J. N. Molecular biology of methanogens. Annu Rev Microbiol. 1992;46:165–191. doi: 10.1146/annurev.mi.46.100192.001121. [DOI] [PubMed] [Google Scholar]
- Robson R. L. Identification of possible adenine nucleotide-binding sites in nitrogenase Fe- and MoFe-proteins by amino acid sequence comparison. FEBS Lett. 1984 Aug 6;173(2):394–398. doi: 10.1016/0014-5793(84)80812-1. [DOI] [PubMed] [Google Scholar]
- Robson R. L., Woodley P. R., Pau R. N., Eady R. R. Structural genes for the vanadium nitrogenase from Azotobacter chroococcum. EMBO J. 1989 Apr;8(4):1217–1224. doi: 10.1002/j.1460-2075.1989.tb03495.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F. Determination of nucleotide sequences in DNA. Science. 1981 Dec 11;214(4526):1205–1210. doi: 10.1126/science.7302589. [DOI] [PubMed] [Google Scholar]
- Sibold L., Henriquet M., Possot O., Aubert J. P. Nucleotide sequence of nifH regions from Methanobacterium ivanovii and Methanosarcina barkeri 227 and characterization of glnB-like genes. Res Microbiol. 1991 Jan;142(1):5–12. doi: 10.1016/0923-2508(91)90091-n. [DOI] [PubMed] [Google Scholar]
- Souillard N., Magot M., Possot O., Sibold L. Nucleotide sequence of regions homologous to nifH (nitrogenase Fe protein) from the nitrogen-fixing archaebacteria Methanococcus thermolithotrophicus and Methanobacterium ivanovii: evolutionary implications. J Mol Evol. 1988;27(1):65–76. doi: 10.1007/BF02099731. [DOI] [PubMed] [Google Scholar]
- Souillard N., Sibold L. Primary structure, functional organization and expression of nitrogenase structural genes of the thermophilic archaebacterium Methanococcus thermolithotrophicus. Mol Microbiol. 1989 Apr;3(4):541–551. doi: 10.1111/j.1365-2958.1989.tb00200.x. [DOI] [PubMed] [Google Scholar]
- Terlesky K. C., Ferry J. G. Ferredoxin requirement for electron transport from the carbon monoxide dehydrogenase complex to a membrane-bound hydrogenase in acetate-grown Methanosarcina thermophila. J Biol Chem. 1988 Mar 25;263(9):4075–4079. [PubMed] [Google Scholar]
- Thiel T. Characterization of genes for an alternative nitrogenase in the cyanobacterium Anabaena variabilis. J Bacteriol. 1993 Oct;175(19):6276–6286. doi: 10.1128/jb.175.19.6276-6286.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiboni O., Cammarano P., Sanangelantoni A. M. Cloning and sequencing of the gene encoding glutamine synthetase I from the archaeum Pyrococcus woesei: anomalous phylogenies inferred from analysis of archaeal and bacterial glutamine synthetase I sequences. J Bacteriol. 1993 May;175(10):2961–2969. doi: 10.1128/jb.175.10.2961-2969.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S. Z., Chen J. S., Johnson J. L. The presence of five nifH-like sequences in Clostridium pasteurianum: sequence divergence and transcription properties. Nucleic Acids Res. 1988 Jan 25;16(2):439–454. doi: 10.1093/nar/16.2.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weston M. F., Kotake S., Davis L. C. Interaction of nitrogenase with nucleotide analogs of ATP and ADP and the effect of metal ions on ADP inhibition. Arch Biochem Biophys. 1983 Sep;225(2):809–817. doi: 10.1016/0003-9861(83)90093-0. [DOI] [PubMed] [Google Scholar]
- Woese C. R. Bacterial evolution. Microbiol Rev. 1987 Jun;51(2):221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinoni F., Robson R. M., Robson R. L. Organization of potential alternative nitrogenase genes from Clostridium pasteurianum. Biochim Biophys Acta. 1993 Jul 18;1174(1):83–86. doi: 10.1016/0167-4781(93)90096-v. [DOI] [PubMed] [Google Scholar]