Abstract
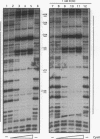
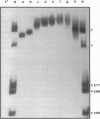
The cynTSX operon enables Escherichia coli K-12 to degrade and use cyanate as a sole nitrogen source. The promoter of this operon is positively regulated by cyanate and the CynR protein. CynR, a member of the LysR family of regulatory proteins, binds specifically to a 136-bp DNA fragment containing both the cynR and the cynTSX promoters. In this study, we report the results of DNase I digestion studies showing that CynR protects a 60-bp region on the cynR coding strand and a 56-bp sequence on the cynTSX coding strand. CynR binding was not affected by cyanate or its structural homolog azide, a gratuitous inducer of the operon. However, CynR-induced bending of two different DNA fragments was detected. The amount of bending was decreased by cyanate.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson P. M. Purification and properties of the inducible enzyme cyanase. Biochemistry. 1980 Jun 24;19(13):2882–2888. doi: 10.1021/bi00554a010. [DOI] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Byerly K. A., Urbanowski M. L., Stauffer G. V. The metR binding site in the Salmonella typhimurium metH gene: DNA sequence constraints on activation. J Bacteriol. 1991 Jun;173(11):3547–3553. doi: 10.1128/jb.173.11.3547-3553.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang M., Crawford I. P. The roles of indoleglycerol phosphate and the TrpI protein in the expression of trpBA from Pseudomonas aeruginosa. Nucleic Acids Res. 1990 Feb 25;18(4):979–988. doi: 10.1093/nar/18.4.979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collado-Vides J., Magasanik B., Gralla J. D. Control site location and transcriptional regulation in Escherichia coli. Microbiol Rev. 1991 Sep;55(3):371–394. doi: 10.1128/mr.55.3.371-394.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher R. F., Long S. R. DNA footprint analysis of the transcriptional activator proteins NodD1 and NodD3 on inducible nod gene promoters. J Bacteriol. 1989 Oct;171(10):5492–5502. doi: 10.1128/jb.171.10.5492-5502.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fried M., Crothers D. M. Equilibria and kinetics of lac repressor-operator interactions by polyacrylamide gel electrophoresis. Nucleic Acids Res. 1981 Dec 11;9(23):6505–6525. doi: 10.1093/nar/9.23.6505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galas D. J., Schmitz A. DNAse footprinting: a simple method for the detection of protein-DNA binding specificity. Nucleic Acids Res. 1978 Sep;5(9):3157–3170. doi: 10.1093/nar/5.9.3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilloton M. B., Lamblin A. F., Kozliak E. I., Gerami-Nejad M., Tu C., Silverman D., Anderson P. M., Fuchs J. A. A physiological role for cyanate-induced carbonic anhydrase in Escherichia coli. J Bacteriol. 1993 Mar;175(5):1443–1451. doi: 10.1128/jb.175.5.1443-1451.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S., Haughn G. W., Calvo J. M., Wallace J. C. A large family of bacterial activator proteins. Proc Natl Acad Sci U S A. 1988 Sep;85(18):6602–6606. doi: 10.1073/pnas.85.18.6602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J. Z., Schell M. A. In vivo interactions of the NahR transcriptional activator with its target sequences. Inducer-mediated changes resulting in transcription activation. J Biol Chem. 1991 Jun 15;266(17):10830–10838. [PubMed] [Google Scholar]
- Kunkel T. A., Roberts J. D., Zakour R. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- Kusano T., Sugawara K. Specific binding of Thiobacillus ferrooxidans RbcR to the intergenic sequence between the rbc operon and the rbcR gene. J Bacteriol. 1993 Feb;175(4):1019–1025. doi: 10.1128/jb.175.4.1019-1025.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LURIA S. E., BURROUS J. W. Hybridization between Escherichia coli and Shigella. J Bacteriol. 1957 Oct;74(4):461–476. doi: 10.1128/jb.74.4.461-476.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamblin A. F., Fuchs J. A. Expression and purification of the cynR regulatory gene product: CynR is a DNA-binding protein. J Bacteriol. 1993 Dec;175(24):7990–7999. doi: 10.1128/jb.175.24.7990-7999.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen J. E., Albrechtsen B., Valentin-Hansen P. Analysis of the terminator region after the deoCABD operon of Escherichia coli K-12 using a new class of single copy number operon-fusion vectors. Nucleic Acids Res. 1987 Jul 10;15(13):5125–5140. doi: 10.1093/nar/15.13.5125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindquist S., Lindberg F., Normark S. Binding of the Citrobacter freundii AmpR regulator to a single DNA site provides both autoregulation and activation of the inducible ampC beta-lactamase gene. J Bacteriol. 1989 Jul;171(7):3746–3753. doi: 10.1128/jb.171.7.3746-3753.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mares R., Urbanowski M. L., Stauffer G. V. Regulation of the Salmonella typhimurium metA gene by the metR protein and homocysteine. J Bacteriol. 1992 Jan;174(2):390–397. doi: 10.1128/jb.174.2.390-397.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsek M. R., Shinabarger D. L., Rothmel R. K., Chakrabarty A. M. Roles of CatR and cis,cis-muconate in activation of the catBC operon, which is involved in benzoate degradation in Pseudomonas putida. J Bacteriol. 1992 Dec;174(23):7798–7806. doi: 10.1128/jb.174.23.7798-7806.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Martín J., Espinosa M. Protein-induced bending as a transcriptional switch. Science. 1993 May 7;260(5109):805–807. doi: 10.1126/science.8387228. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schell M. A., Poser E. F. Demonstration, characterization, and mutational analysis of NahR protein binding to nah and sal promoters. J Bacteriol. 1989 Feb;171(2):837–846. doi: 10.1128/jb.171.2.837-846.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz S. C., Shields G. C., Steitz T. A. Crystal structure of a CAP-DNA complex: the DNA is bent by 90 degrees. Science. 1991 Aug 30;253(5023):1001–1007. doi: 10.1126/science.1653449. [DOI] [PubMed] [Google Scholar]
- Simons R. W., Houman F., Kleckner N. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene. 1987;53(1):85–96. doi: 10.1016/0378-1119(87)90095-3. [DOI] [PubMed] [Google Scholar]
- Sung Y. C., Fuchs J. A. Characterization of the cyn operon in Escherichia coli K12. J Biol Chem. 1988 Oct 15;263(29):14769–14775. [PubMed] [Google Scholar]
- Sung Y. C., Fuchs J. A. The Escherichia coli K-12 cyn operon is positively regulated by a member of the lysR family. J Bacteriol. 1992 Jun;174(11):3645–3650. doi: 10.1128/jb.174.11.3645-3650.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung Y. C., Parsell D., Anderson P. M., Fuchs J. A. Identification, mapping, and cloning of the gene encoding cyanase in Escherichia coli K-12. J Bacteriol. 1987 Jun;169(6):2639–2642. doi: 10.1128/jb.169.6.2639-2642.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas M. S., Glass R. E. Escherichia coli rpoA mutation which impairs transcription of positively regulated systems. Mol Microbiol. 1991 Nov;5(11):2719–2725. doi: 10.1111/j.1365-2958.1991.tb01980.x. [DOI] [PubMed] [Google Scholar]
- Thompson J. F., Landy A. Empirical estimation of protein-induced DNA bending angles: applications to lambda site-specific recombination complexes. Nucleic Acids Res. 1988 Oct 25;16(20):9687–9705. doi: 10.1093/nar/16.20.9687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira J., Messing J. Production of single-stranded plasmid DNA. Methods Enzymol. 1987;153:3–11. doi: 10.1016/0076-6879(87)53044-0. [DOI] [PubMed] [Google Scholar]
- Wang L., Helmann J. D., Winans S. C. The A. tumefaciens transcriptional activator OccR causes a bend at a target promoter, which is partially relaxed by a plant tumor metabolite. Cell. 1992 May 15;69(4):659–667. doi: 10.1016/0092-8674(92)90229-6. [DOI] [PubMed] [Google Scholar]
- Wek R. C., Hatfield G. W. Transcriptional activation at adjacent operators in the divergent-overlapping ilvY and ilvC promoters of Escherichia coli. J Mol Biol. 1988 Oct 5;203(3):643–663. doi: 10.1016/0022-2836(88)90199-4. [DOI] [PubMed] [Google Scholar]
- Wu H. M., Crothers D. M. The locus of sequence-directed and protein-induced DNA bending. Nature. 1984 Apr 5;308(5959):509–513. doi: 10.1038/308509a0. [DOI] [PubMed] [Google Scholar]
- Zinkel S. S., Crothers D. M. Catabolite activator protein-induced DNA bending in transcription initiation. J Mol Biol. 1991 May 20;219(2):201–215. doi: 10.1016/0022-2836(91)90562-k. [DOI] [PubMed] [Google Scholar]
- Zwieb C., Brown R. S. Absence of substantial bending in Xenopus laevis transcription factor IIIA-DNA complexes. Nucleic Acids Res. 1990 Feb 11;18(3):583–587. doi: 10.1093/nar/18.3.583. [DOI] [PMC free article] [PubMed] [Google Scholar]