Abstract
The mucoid phenotype is common among strains of Pseudomonas aeruginosa that cause chronic pulmonary infections in patients with cystic fibrosis and is due to overproduction of an exopolysaccharide called alginate. However, the mucoid phenotype is unstable in vitro, especially when the cells are incubated under low oxygen tension. Spontaneous conversion to the nonmucoid form is typically due to mutations (previously called algS) that are closely linked to the alginate regulatory gene algT, located at 68 min on the chromosome. Our sequence analysis of algT showed that its 22-kDa gene product shares homology with several alternate sigma factors in bacteria, suggesting that AlgT (also known as AlgU) interacts directly with RNA polymerase core to activate the promoters of alginate genes. AlgT showed striking sequence similarity (79%) to sigma E of Escherichia coli, an alternate sigma factor involved in high-temperature gene expression. Our analysis of the molecular basis for spontaneous conversion from mucoid to nonmucoid, in the cystic fibrosis isolate FRD, revealed that nonmucoid conversion was often due to one of two distinct missense mutations in algT that occurred at codons 18 and 29. RNase protection assays showed that spontaneous nonmucoid strains with the algT18 and algT29 alleles have a four- to fivefold reduction in the accumulation of algT transcripts compared with the wild-type mucoid strain. Likewise, a plasmid-borne algT-cat transcriptional fusion was about 3-fold less active in the algT18 and algT29 backgrounds compared with the mucoid wild-type strain, and it was 20-fold less active in an algT::Tn501 background. These data indicate that algT is autoregulated. The spontaneous algT missense alleles also caused about fivefold-reduced expression of the adjacent negative regulator, algN (also known as mucB). Transcripts of algN were essentially absent in the algT::Tn501 strain. Thus, algT regulates the algTN cluster, and the two genes may be cotranscribed. A primer extension analysis showed that algT transcription starts 54 bp upstream of the start of translation. Although the algT promoter showed little similarity to promoters recognized by the vegetative sigma factor, it was similar to the algR promoter. This finding suggests that AlgT may function as a sigma factor to activate its own promoter and those of other alginate genes. The primer extension analysis also showed that algT transcripts were readily detectable in the typical nonmucoid strain PAO1, which was in contrast to a weak signal seen in the algT18 mutant of FRD. A plasmid-borne algT gene in PAO1 resulted in both the mucoid phenotype and high levels of algT transcripts, further supporting the hypothesis that AlgT controls its own gene expression and expression of genes of the alginate regulon.
Full text
PDF









Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. Basic local alignment search tool. J Mol Biol. 1990 Oct 5;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Baltimore R. S., Mitchell M. Immunologic investigations of mucoid strains of Pseudomonas aeruginosa: comparison of susceptibility to opsonic antibody in mucoid and nonmucoid strains. J Infect Dis. 1980 Feb;141(2):238–247. doi: 10.1093/infdis/141.2.238. [DOI] [PubMed] [Google Scholar]
- Benson A. K., Haldenwang W. G. Bacillus subtilis sigma B is regulated by a binding protein (RsbW) that blocks its association with core RNA polymerase. Proc Natl Acad Sci U S A. 1993 Mar 15;90(6):2330–2334. doi: 10.1073/pnas.90.6.2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brumlik M. J., Storey D. G. Zinc and iron regulate translation of the gene encoding Pseudomonas aeruginosa elastase. Mol Microbiol. 1992 Feb;6(3):337–344. doi: 10.1111/j.1365-2958.1992.tb01476.x. [DOI] [PubMed] [Google Scholar]
- Chitnis C. E., Ohman D. E. Genetic analysis of the alginate biosynthetic gene cluster of Pseudomonas aeruginosa shows evidence of an operonic structure. Mol Microbiol. 1993 May;8(3):583–593. doi: 10.1111/j.1365-2958.1993.tb01602.x. [DOI] [PubMed] [Google Scholar]
- Close T. J., Rodriguez R. L. Construction and characterization of the chloramphenicol-resistance gene cartridge: a new approach to the transcriptional mapping of extrachromosomal elements. Gene. 1982 Dec;20(2):305–316. doi: 10.1016/0378-1119(82)90048-8. [DOI] [PubMed] [Google Scholar]
- Darzins A., Chakrabarty A. M. Cloning of genes controlling alginate biosynthesis from a mucoid cystic fibrosis isolate of Pseudomonas aeruginosa. J Bacteriol. 1984 Jul;159(1):9–18. doi: 10.1128/jb.159.1.9-18.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deretic V., Dikshit R., Konyecsni W. M., Chakrabarty A. M., Misra T. K. The algR gene, which regulates mucoidy in Pseudomonas aeruginosa, belongs to a class of environmentally responsive genes. J Bacteriol. 1989 Mar;171(3):1278–1283. doi: 10.1128/jb.171.3.1278-1283.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deretic V., Gill J. F., Chakrabarty A. M. Pseudomonas aeruginosa infection in cystic fibrosis: nucleotide sequence and transcriptional regulation of the algD gene. Nucleic Acids Res. 1987 Jun 11;15(11):4567–4581. doi: 10.1093/nar/15.11.4567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deretic V., Schurr M. J., Boucher J. C., Martin D. W. Conversion of Pseudomonas aeruginosa to mucoidy in cystic fibrosis: environmental stress and regulation of bacterial virulence by alternative sigma factors. J Bacteriol. 1994 May;176(10):2773–2780. doi: 10.1128/jb.176.10.2773-2780.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubnau E., Weir J., Nair G., Carter L., 3rd, Moran C., Jr, Smith I. Bacillus sporulation gene spo0H codes for sigma 30 (sigma H). J Bacteriol. 1988 Mar;170(3):1054–1062. doi: 10.1128/jb.170.3.1054-1062.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson J. W., Gross C. A. Identification of the sigma E subunit of Escherichia coli RNA polymerase: a second alternate sigma factor involved in high-temperature gene expression. Genes Dev. 1989 Sep;3(9):1462–1471. doi: 10.1101/gad.3.9.1462. [DOI] [PubMed] [Google Scholar]
- Evans L. R., Linker A. Production and characterization of the slime polysaccharide of Pseudomonas aeruginosa. J Bacteriol. 1973 Nov;116(2):915–924. doi: 10.1128/jb.116.2.915-924.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figurski D. H., Helinski D. R. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn J. L., Ohman D. E. Cloning of genes from mucoid Pseudomonas aeruginosa which control spontaneous conversion to the alginate production phenotype. J Bacteriol. 1988 Apr;170(4):1452–1460. doi: 10.1128/jb.170.4.1452-1460.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn J. L., Ohman D. E. Use of a gene replacement cosmid vector for cloning alginate conversion genes from mucoid and nonmucoid Pseudomonas aeruginosa strains: algS controls expression of algT. J Bacteriol. 1988 Jul;170(7):3228–3236. doi: 10.1128/jb.170.7.3228-3236.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin M. J., Chitnis C. E., Gacesa P., Sonesson A., White D. C., Ohman D. E. Pseudomonas aeruginosa AlgG is a polymer level alginate C5-mannuronan epimerase. J Bacteriol. 1994 Apr;176(7):1821–1830. doi: 10.1128/jb.176.7.1821-1830.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin M. J., Ohman D. E. Identification of algF in the alginate biosynthetic gene cluster of Pseudomonas aeruginosa which is required for alginate acetylation. J Bacteriol. 1993 Aug;175(16):5057–5065. doi: 10.1128/jb.175.16.5057-5065.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freitag C. S., Abraham J. M., Clements J. R., Eisenstein B. I. Genetic analysis of the phase variation control of expression of type 1 fimbriae in Escherichia coli. J Bacteriol. 1985 May;162(2):668–675. doi: 10.1128/jb.162.2.668-675.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman A. M., Long S. R., Brown S. E., Buikema W. J., Ausubel F. M. Construction of a broad host range cosmid cloning vector and its use in the genetic analysis of Rhizobium mutants. Gene. 1982 Jun;18(3):289–296. doi: 10.1016/0378-1119(82)90167-6. [DOI] [PubMed] [Google Scholar]
- Fyfe J. A., Govan J. R. Alginate synthesis in mucoid Pseudomonas aeruginosa: a chromosomal locus involved in control. J Gen Microbiol. 1980 Aug;119(2):443–450. doi: 10.1099/00221287-119-2-443. [DOI] [PubMed] [Google Scholar]
- Goldberg J. B., Dahnke T. Pseudomonas aeruginosa AlgB, which modulates the expression of alginate, is a member of the NtrC subclass of prokaryotic regulators. Mol Microbiol. 1992 Jan;6(1):59–66. doi: 10.1111/j.1365-2958.1992.tb00837.x. [DOI] [PubMed] [Google Scholar]
- Goldberg J. B., Gorman W. L., Flynn J. L., Ohman D. E. A mutation in algN permits trans activation of alginate production by algT in Pseudomonas species. J Bacteriol. 1993 Mar;175(5):1303–1308. doi: 10.1128/jb.175.5.1303-1308.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg J. B., Ohman D. E. Cloning and expression in Pseudomonas aeruginosa of a gene involved in the production of alginate. J Bacteriol. 1984 Jun;158(3):1115–1121. doi: 10.1128/jb.158.3.1115-1121.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govan J. R., Fyfe J. A., Jarman T. R. Isolation of alginate-producing mutants of Pseudomonas fluorescens, Pseudomonas putida and Pseudomonas mendocina. J Gen Microbiol. 1981 Jul;125(1):217–220. doi: 10.1099/00221287-125-1-217. [DOI] [PubMed] [Google Scholar]
- Govan J. R., Fyfe J. A. Mucoid Pseudomonas aeruginosa and cystic fibrosis: resistance of the mucoid from to carbenicillin, flucloxacillin and tobramycin and the isolation of mucoid variants in vitro. J Antimicrob Chemother. 1978 May;4(3):233–240. doi: 10.1093/jac/4.3.233. [DOI] [PubMed] [Google Scholar]
- Govan J. R., Harris G. S. Pseudomonas aeruginosa and cystic fibrosis: unusual bacterial adaptation and pathogenesis. Microbiol Sci. 1986 Oct;3(10):302–308. [PubMed] [Google Scholar]
- Haas D., Holloway B. W. R factor variants with enhanced sex factor activity in Pseudomonas aeruginosa. Mol Gen Genet. 1976 Mar 30;144(3):243–251. doi: 10.1007/BF00341722. [DOI] [PubMed] [Google Scholar]
- Hancock R. E., Mutharia L. M., Chan L., Darveau R. P., Speert D. P., Pier G. B. Pseudomonas aeruginosa isolates from patients with cystic fibrosis: a class of serum-sensitive, nontypable strains deficient in lipopolysaccharide O side chains. Infect Immun. 1983 Oct;42(1):170–177. doi: 10.1128/iai.42.1.170-177.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holloway B. W., Morgan A. F. Genome organization in Pseudomonas. Annu Rev Microbiol. 1986;40:79–105. doi: 10.1146/annurev.mi.40.100186.000455. [DOI] [PubMed] [Google Scholar]
- Kimbara K., Chakrabarty A. M. Control of alginate synthesis in Pseudomonas aeruginosa: regulation of the algR1 gene. Biochem Biophys Res Commun. 1989 Oct 31;164(2):601–608. doi: 10.1016/0006-291x(89)91502-7. [DOI] [PubMed] [Google Scholar]
- Lam J., Chan R., Lam K., Costerton J. W. Production of mucoid microcolonies by Pseudomonas aeruginosa within infected lungs in cystic fibrosis. Infect Immun. 1980 May;28(2):546–556. doi: 10.1128/iai.28.2.546-556.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindgren P. B., Frederick R., Govindarajan A. G., Panopoulos N. J., Staskawicz B. J., Lindow S. E. An ice nucleation reporter gene system: identification of inducible pathogenicity genes in Pseudomonas syringae pv. phaseolicola. EMBO J. 1989 May;8(5):1291–1301. doi: 10.1002/j.1460-2075.1989.tb03508.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonetto M., Gribskov M., Gross C. A. The sigma 70 family: sequence conservation and evolutionary relationships. J Bacteriol. 1992 Jun;174(12):3843–3849. doi: 10.1128/jb.174.12.3843-3849.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luzar M. A., Montie T. C. Avirulence and altered physiological properties of cystic fibrosis strains of Pseudomonas aeruginosa. Infect Immun. 1985 Nov;50(2):572–576. doi: 10.1128/iai.50.2.572-576.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin D. W., Holloway B. W., Deretic V. Characterization of a locus determining the mucoid status of Pseudomonas aeruginosa: AlgU shows sequence similarities with a Bacillus sigma factor. J Bacteriol. 1993 Feb;175(4):1153–1164. doi: 10.1128/jb.175.4.1153-1164.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin D. W., Schurr M. J., Mudd M. H., Govan J. R., Holloway B. W., Deretic V. Mechanism of conversion to mucoidy in Pseudomonas aeruginosa infecting cystic fibrosis patients. Proc Natl Acad Sci U S A. 1993 Sep 15;90(18):8377–8381. doi: 10.1073/pnas.90.18.8377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May T. B., Shinabarger D., Maharaj R., Kato J., Chu L., DeVault J. D., Roychoudhury S., Zielinski N. A., Berry A., Rothmel R. K. Alginate synthesis by Pseudomonas aeruginosa: a key pathogenic factor in chronic pulmonary infections of cystic fibrosis patients. Clin Microbiol Rev. 1991 Apr;4(2):191–206. doi: 10.1128/cmr.4.2.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mian F. A., Jarman T. R., Righelato R. C. Biosynthesis of exopolysaccharide by Pseudomonas aeruginosa. J Bacteriol. 1978 May;134(2):418–422. doi: 10.1128/jb.134.2.418-422.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J. F., Mekalanos J. J., Falkow S. Coordinate regulation and sensory transduction in the control of bacterial virulence. Science. 1989 Feb 17;243(4893):916–922. doi: 10.1126/science.2537530. [DOI] [PubMed] [Google Scholar]
- Ogle J. W., Janda J. M., Woods D. E., Vasil M. L. Characterization and use of a DNA probe as an epidemiological marker for Pseudomonas aeruginosa. J Infect Dis. 1987 Jan;155(1):119–126. doi: 10.1093/infdis/155.1.119. [DOI] [PubMed] [Google Scholar]
- Ohman D. E., Chakrabarty A. M. Genetic mapping of chromosomal determinants for the production of the exopolysaccharide alginate in a Pseudomonas aeruginosa cystic fibrosis isolate. Infect Immun. 1981 Jul;33(1):142–148. doi: 10.1128/iai.33.1.142-148.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohman D. E., Chakrabarty A. M. Utilization of human respiratory secretions by mucoid Pseudomonas aeruginosa of cystic fibrosis origin. Infect Immun. 1982 Aug;37(2):662–669. doi: 10.1128/iai.37.2.662-669.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohman D. E., West M. A., Flynn J. L., Goldberg J. B. Method for gene replacement in Pseudomonas aeruginosa used in construction of recA mutants: recA-independent instability of alginate production. J Bacteriol. 1985 Jun;162(3):1068–1074. doi: 10.1128/jb.162.3.1068-1074.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson B. D., Meyer T. F. Genetic variation in pathogenic bacteria. Trends Genet. 1992 Dec;8(12):422–427. doi: 10.1016/0168-9525(92)90325-x. [DOI] [PubMed] [Google Scholar]
- Schiller N. L., Monday S. R., Boyd C. M., Keen N. T., Ohman D. E. Characterization of the Pseudomonas aeruginosa alginate lyase gene (algL): cloning, sequencing, and expression in Escherichia coli. J Bacteriol. 1993 Aug;175(15):4780–4789. doi: 10.1128/jb.175.15.4780-4789.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VOGEL H. J., BONNER D. M. Acetylornithinase of Escherichia coli: partial purification and some properties. J Biol Chem. 1956 Jan;218(1):97–106. [PubMed] [Google Scholar]
- Wise K. S. Adaptive surface variation in mycoplasmas. Trends Microbiol. 1993 May;1(2):59–63. doi: 10.1016/0966-842X(93)90034-O. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods D. E., Sokol P. A., Bryan L. E., Storey D. G., Mattingly S. J., Vogel H. J., Ceri H. In vivo regulation of virulence in Pseudomonas aeruginosa associated with genetic rearrangement. J Infect Dis. 1991 Jan;163(1):143–149. doi: 10.1093/infdis/163.1.143. [DOI] [PubMed] [Google Scholar]
- Wozniak D. J., Ohman D. E. Involvement of the alginate algT gene and integration host factor in the regulation of the Pseudomonas aeruginosa algB gene. J Bacteriol. 1993 Jul;175(13):4145–4153. doi: 10.1128/jb.175.13.4145-4153.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wozniak D. J., Ohman D. E. Pseudomonas aeruginosa AlgB, a two-component response regulator of the NtrC family, is required for algD transcription. J Bacteriol. 1991 Feb;173(4):1406–1413. doi: 10.1128/jb.173.4.1406-1413.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
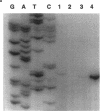
- Wozniak D. J., Ohman D. E. Transcriptional analysis of the Pseudomonas aeruginosa genes algR, algB, and algD reveals a hierarchy of alginate gene expression which is modulated by algT. J Bacteriol. 1994 Oct;176(19):6007–6014. doi: 10.1128/jb.176.19.6007-6014.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y., Heu S., Yi J., Lu Y., Hutcheson S. W. Identification of a putative alternate sigma factor and characterization of a multicomponent regulatory cascade controlling the expression of Pseudomonas syringae pv. syringae Pss61 hrp and hrmA genes. J Bacteriol. 1994 Feb;176(4):1025–1036. doi: 10.1128/jb.176.4.1025-1036.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]