Abstract
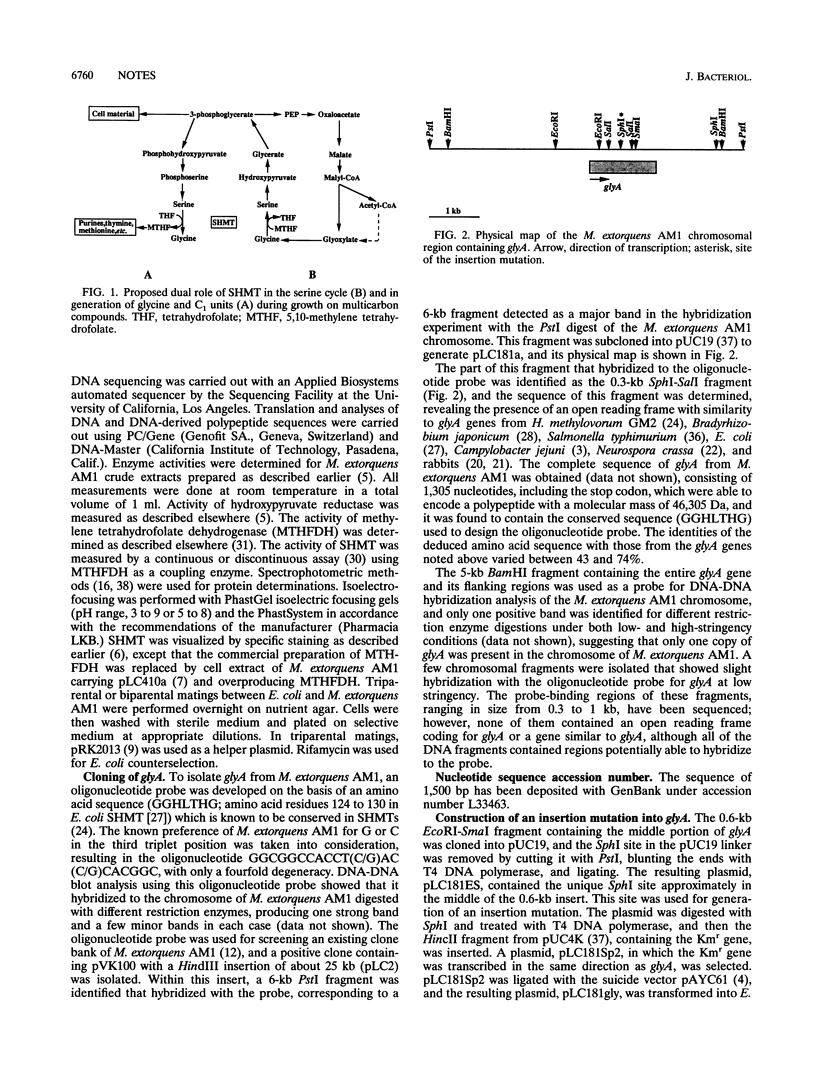
The gene (glyA) of Methylobacterium extorquens AM1 encoding serine hydroxymethyltransferase (SHMT), one of the key enzymes of the serine cycle for C1 assimilation, was isolated by using a synthetic oligonucleotide with a sequence based on amino acid sequence conserved in SHMTs from different sources. The amino acid sequence deduced from the gene revealed high similarity to those of known SHMTs. The cloned gene was inactivated by insertion of a kanamycin resistance gene, and recombination of this insertion derivative with the wild-type gene produced an SHMT null mutant. Surprisingly, this mutant had lost its ability to grow on C1 as well as on C2 compounds but was still able to grow on succinate. The DNA fragment containing glyA was shown not to be linked with fragments carrying serine cycle genes identified earlier, making it the fourth chromosomal region of M. extorquens AM1 to be indicated as being involved in C1 assimilation.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arps P. J., Fulton G. F., Minnich E. C., Lidstrom M. E. Genetics of serine pathway enzymes in Methylobacterium extorquens AM1: phosphoenolpyruvate carboxylase and malyl coenzyme A lyase. J Bacteriol. 1993 Jun;175(12):3776–3783. doi: 10.1128/jb.175.12.3776-3783.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan V. L., Bingham H. L. Complete sequence of the Campylobacter jejuni glyA gene encoding serine hydroxymethyltransferase. Gene. 1991 May 15;101(1):51–58. doi: 10.1016/0378-1119(91)90223-x. [DOI] [PubMed] [Google Scholar]
- Chistoserdov A. Y., Chistoserdova L. V., McIntire W. S., Lidstrom M. E. Genetic organization of the mau gene cluster in Methylobacterium extorquens AM1: complete nucleotide sequence and generation and characteristics of mau mutants. J Bacteriol. 1994 Jul;176(13):4052–4065. doi: 10.1128/jb.176.13.4052-4065.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chistoserdova L. V., Lidstrom M. E. Cloning, mutagenesis, and physiological effect of a hydroxypyruvate reductase gene from Methylobacterium extorquens AM1. J Bacteriol. 1992 Jan;174(1):71–77. doi: 10.1128/jb.174.1.71-77.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chistoserdova L. V., Lidstrom M. E. Genetics of the serine cycle in Methylobacterium extorquens AM1: identification of sgaA and mtdA and sequences of sgaA, hprA, and mtdA. J Bacteriol. 1994 Apr;176(7):1957–1968. doi: 10.1128/jb.176.7.1957-1968.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chistoserdova L. V., Lidstrom M. E. Purification and characterization of hydroxypyruvate reductase from the facultative methylotroph Methylobacterium extorquens AM1. J Bacteriol. 1991 Nov;173(22):7228–7232. doi: 10.1128/jb.173.22.7228-7232.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditta G., Schmidhauser T., Yakobson E., Lu P., Liang X. W., Finlay D. R., Guiney D., Helinski D. R. Plasmids related to the broad host range vector, pRK290, useful for gene cloning and for monitoring gene expression. Plasmid. 1985 Mar;13(2):149–153. doi: 10.1016/0147-619x(85)90068-x. [DOI] [PubMed] [Google Scholar]
- Dunstan P. M., Anthony C., Drabble W. T. Microbial metabolism of C 1 and C 2 compounds. The involvement of glycollate in the metabolism of ethanol and of acetate by Pseudomonas AM1. Biochem J. 1972 Jun;128(1):99–106. doi: 10.1042/bj1280099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunstan P. M., Anthony C., Drabble W. T. Microbial metabolism of C 1 and C 2 compounds. The role of glyoxylate, glycollate and acetate in the growth of Pseudomonas AM1 on ethanol and on C 1 compounds. Biochem J. 1972 Jun;128(1):107–115. doi: 10.1042/bj1280107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulton G. L., Nunn D. N., Lidstrom M. E. Molecular cloning of a malyl coenzyme A lyase gene from Pseudomonas sp. strain AM1, a facultative methylotroph. J Bacteriol. 1984 Nov;160(2):718–723. doi: 10.1128/jb.160.2.718-723.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harder W., Quayle J. R. The biosynthesis of serine and glycine in Pseudomonas AM1 with special reference to growth on carbon sources other than C1 compounds. Biochem J. 1971 Mar;121(5):753–762. doi: 10.1042/bj1210753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heptinstall J., Quayle J. R. Pathways leading to and from serine during growth of Pseudomonas AM1 on C1 compounds or succinate. Biochem J. 1970 Apr;117(3):563–572. doi: 10.1042/bj1170563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalb V. F., Jr, Bernlohr R. W. A new spectrophotometric assay for protein in cell extracts. Anal Biochem. 1977 Oct;82(2):362–371. doi: 10.1016/0003-2697(77)90173-7. [DOI] [PubMed] [Google Scholar]
- Lidstrom M. E., Tsygankov Y. D. Molecular genetics of methylotrophic bacteria. Biotechnology. 1991;18:273–304. doi: 10.1016/b978-0-7506-9188-8.50019-x. [DOI] [PubMed] [Google Scholar]
- Martini F., Angelaccio S., Pascarella S., Barra D., Bossa F., Schirch V. The primary structure of rabbit liver cytosolic serine hydroxymethyltransferase. J Biol Chem. 1987 Apr 25;262(12):5499–5509. [PubMed] [Google Scholar]
- Martini F., Maras B., Tanci P., Angelaccio S., Pascarella S., Barra D., Bossa F., Schirch V. The primary structure of rabbit liver mitochondrial serine hydroxymethyltransferase. J Biol Chem. 1989 May 25;264(15):8509–8519. [PubMed] [Google Scholar]
- McClung C. R., Davis C. R., Page K. M., Denome S. A. Characterization of the formate (for) locus, which encodes the cytosolic serine hydroxymethyltransferase of Neurospora crassa. Mol Cell Biol. 1992 Apr;12(4):1412–1421. doi: 10.1128/mcb.12.4.1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinkoth J., Wahl G. Hybridization of nucleic acids immobilized on solid supports. Anal Biochem. 1984 May 1;138(2):267–284. doi: 10.1016/0003-2697(84)90808-x. [DOI] [PubMed] [Google Scholar]
- Miyata A., Yoshida T., Yamaguchi K., Yokoyama C., Tanabe T., Toh H., Mitsunaga T., Izumi Y. Molecular cloning and expression of the gene for serine hydroxymethyltransferase from an obligate methylotroph Hyphomicrobium methylovorum GM2. Eur J Biochem. 1993 Mar 15;212(3):745–750. doi: 10.1111/j.1432-1033.1993.tb17713.x. [DOI] [PubMed] [Google Scholar]
- O'Connor M. L., Hanson R. S. Serine transhydroxymethylase isoenzymes from a facultative methylotroph. J Bacteriol. 1975 Nov;124(2):985–996. doi: 10.1128/jb.124.2.985-996.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PEEL D., QUAYLE J. R. Microbial growth on C1 compounds. I. Isolation and characterization of Pseudomonas AM 1. Biochem J. 1961 Dec;81:465–469. doi: 10.1042/bj0810465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plamann M. D., Stauffer L. T., Urbanowski M. L., Stauffer G. V. Complete nucleotide sequence of the E. coli glyA gene. Nucleic Acids Res. 1983 Apr 11;11(7):2065–2075. doi: 10.1093/nar/11.7.2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossbach S., Hennecke H. Identification of glyA as a symbiotically essential gene in Bradyrhizobium japonicum. Mol Microbiol. 1991 Jan;5(1):39–47. doi: 10.1111/j.1365-2958.1991.tb01824.x. [DOI] [PubMed] [Google Scholar]
- SAITO H., MIURA K. I. PREPARATION OF TRANSFORMING DEOXYRIBONUCLEIC ACID BY PHENOL TREATMENT. Biochim Biophys Acta. 1963 Aug 20;72:619–629. [PubMed] [Google Scholar]
- Tsuji K., Tsien H. C., Hanson R. S., DePalma S. R., Scholtz R., LaRoche S. 16S ribosomal RNA sequence analysis for determination of phylogenetic relationship among methylotrophs. J Gen Microbiol. 1990 Jan;136(1):1–10. doi: 10.1099/00221287-136-1-1. [DOI] [PubMed] [Google Scholar]
- UMBARGER H. E., UMBARGER M. A., SIU P. M. BIOSYNTHESIS OF SERINE IN ESCHERICHIA COLI AND SALMONELLA TYPHIMURIUM. J Bacteriol. 1963 Jun;85:1431–1439. doi: 10.1128/jb.85.6.1431-1439.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbanowski M. L., Plamann M. D., Stauffer L. T., Stauffer G. V. Cloning and characterization of the gene for Salmonella typhimurium serine hydroxymethyltransferase. Gene. 1984 Jan;27(1):47–54. doi: 10.1016/0378-1119(84)90237-3. [DOI] [PubMed] [Google Scholar]
- Vieira J., Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982 Oct;19(3):259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- Whitaker J. R., Granum P. E. An absolute method for protein determination based on difference in absorbance at 235 and 280 nm. Anal Biochem. 1980 Nov 15;109(1):156–159. doi: 10.1016/0003-2697(80)90024-x. [DOI] [PubMed] [Google Scholar]