Abstract
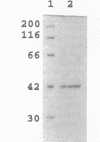
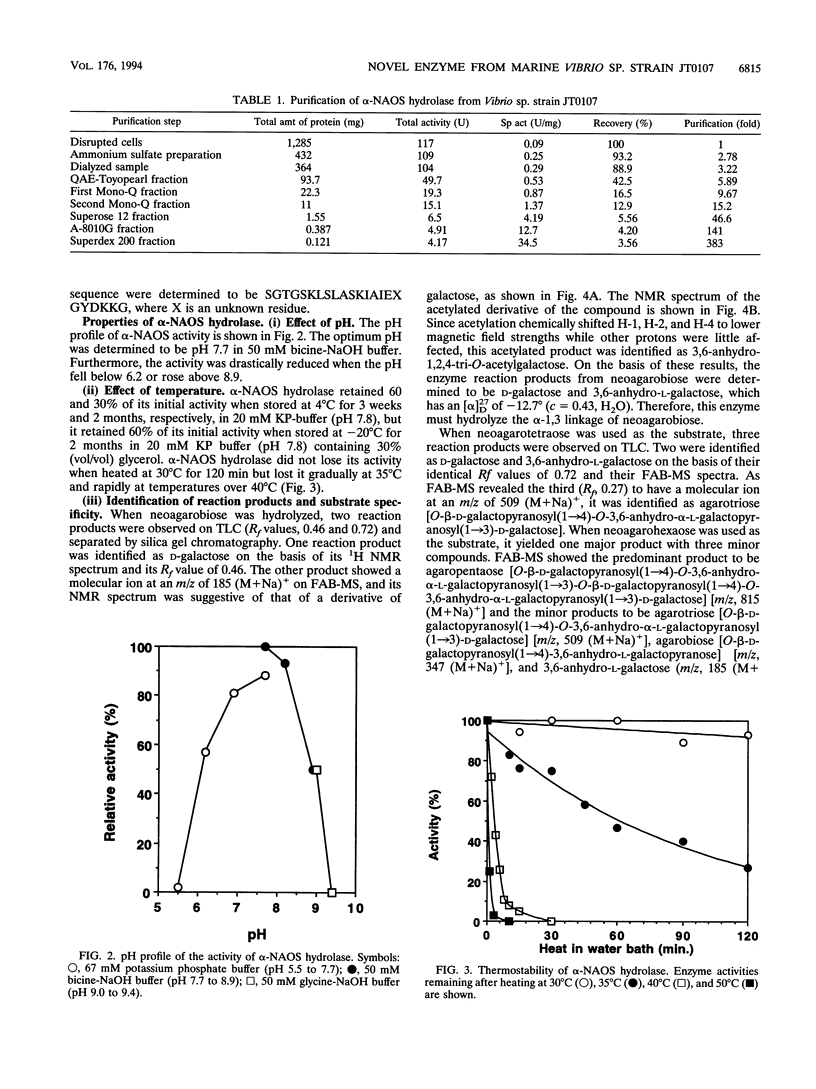
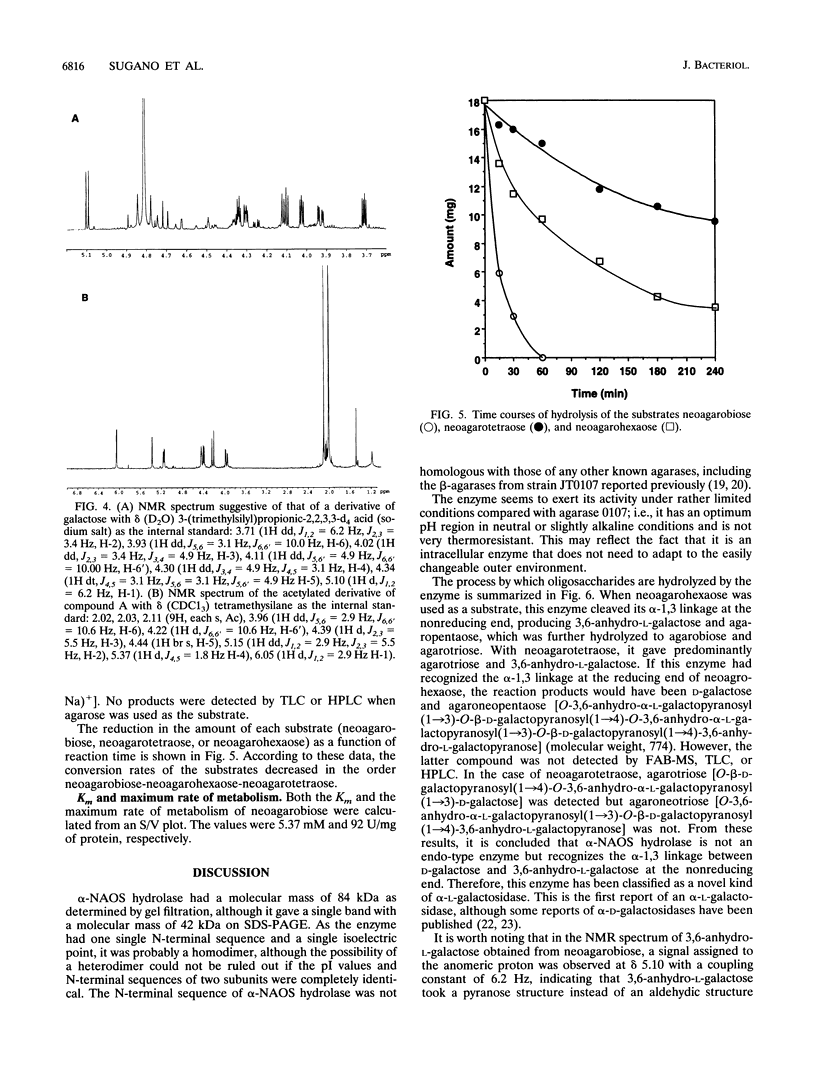
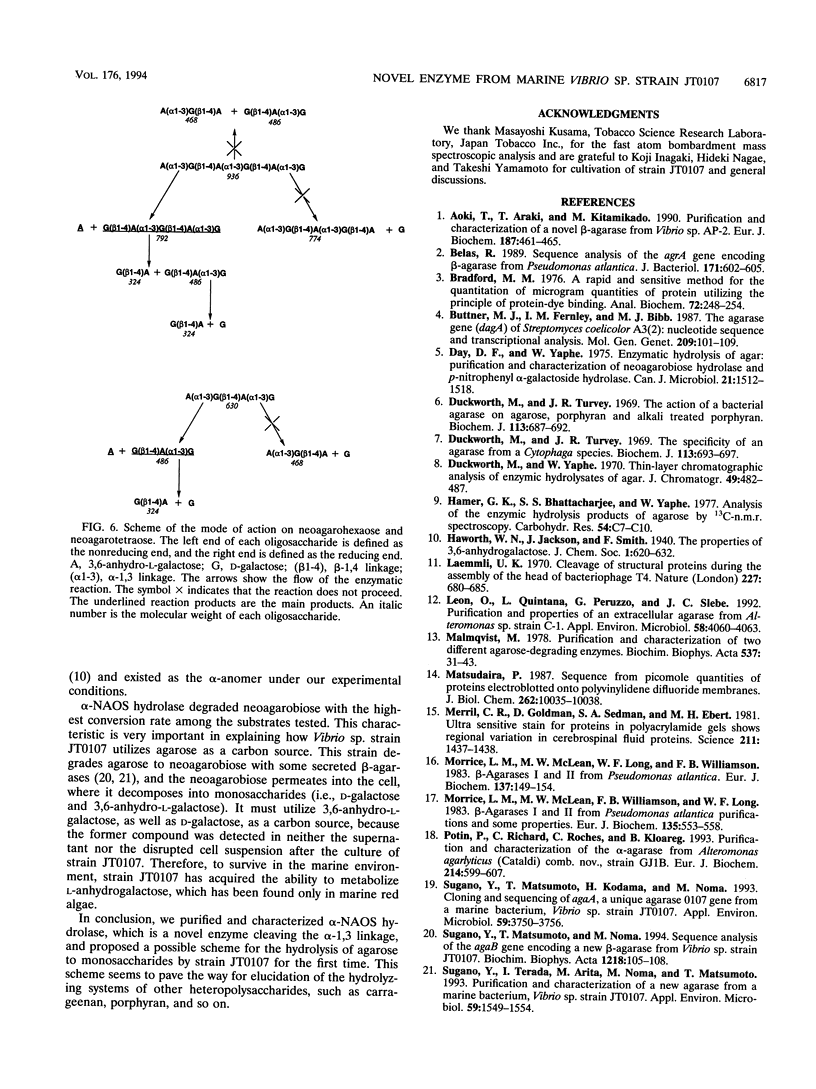
A novel enzyme, alpha-neoagarooligosaccharide hydrolase (EC 3.2.1.-), which hydrolyzes the alpha-1,3 linkage of neoagarooligosaccharides to yield agaropentaose (O-beta-D-galactopyranosyl(1-->4)-O-3,6-anhydro-alpha-L-galactopyranosyl (1-->3)-D-galactose], agarotriose [O-beta-D-galactopyranosyl(1-->4)-O-3,6-anhydro- alpha-L-galactopyranosyl (1-->3)-D-galactose], agarobiose [O-beta-D-galactopyranosyl(1-->4)-3,6-anhydro-L-galactose], 3,6-anhydro-L-galactose, and D-galactose was isolated from the marine bacterium Vibrio sp. strain JT0107 and characterized. This enzyme was purified 383-fold from cultured cells by using a combination of ammonium sulfate precipitation, successive anion-exchange column chromatography, gel filtration, and hydroxyapatite chromatography, gel filtration, and hydroxyapatite chromatography. The purified protein gave a single band (M(r), 42,000) on sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Estimation of the M(r) by the gel filtration method gave a value of 84,000, indicating that the enzyme is dimeric. Amino acid sequence analysis revealed it to have a single N-terminal sequence that has no sequence homology to any other known agarases. The optimum temperature and pH were 30 degrees C and 7.7, respectively. The Km and maximum rate of metabolism for neoagarobiose were 5.37 mM and 92 U/mg of protein, respectively.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aoki T., Araki T., Kitamikado M. Purification and characterization of a novel beta-agarase from Vibrio sp. AP-2. Eur J Biochem. 1990 Jan 26;187(2):461–465. doi: 10.1111/j.1432-1033.1990.tb15326.x. [DOI] [PubMed] [Google Scholar]
- Belas R. Sequence analysis of the agrA gene encoding beta-agarase from Pseudomonas atlantica. J Bacteriol. 1989 Jan;171(1):602–605. doi: 10.1128/jb.171.1.602-605.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Buttner M J, Fearnley I M, Bibb M J. The agarase gene (dagA) of Streptomyces coelicolor A3(2): nucleotide sequence and transcriptional analysis. Mol Gen Genet. 1987 Aug;209(1):101–109. doi: 10.1007/BF00329843. [DOI] [PubMed] [Google Scholar]
- Day D. F., Yaphe W. Enzymatic hydrolysis of agar: purification and characterization of neoagarobiose hydrolase and p-nitrophenyl alpha-galactoside hydrolase. Can J Microbiol. 1975 Oct;21(10):1512–1518. doi: 10.1139/m75-223. [DOI] [PubMed] [Google Scholar]
- Duckworth M., Turvey J. R. The action of a bacterial agarase on agarose, porphyran and alkali-treated porphyran. Biochem J. 1969 Jul;113(4):687–692. doi: 10.1042/bj1130687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duckworth M., Turvey J. R. The specificity of an agarase from a Cytophaga species. Biochem J. 1969 Jul;113(4):693–696. doi: 10.1042/bj1130693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duckworth M., Yaphe W. Thin-layer chromatographic analysis of enzymic hydrolysates of agar. J Chromatogr. 1970 Jun 24;49(3):482–487. doi: 10.1016/s0021-9673(00)93663-x. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Leon O., Quintana L., Peruzzo G., Slebe J. C. Purification and Properties of an Extracellular Agarase from Alteromonas sp. Strain C-1. Appl Environ Microbiol. 1992 Dec;58(12):4060–4063. doi: 10.1128/aem.58.12.4060-4063.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malmqvist M. Purification and characterization of two different agarose-degrading enzymes. Biochim Biophys Acta. 1978 Nov 20;537(1):31–43. doi: 10.1016/0005-2795(78)90600-1. [DOI] [PubMed] [Google Scholar]
- Matsudaira P. Sequence from picomole quantities of proteins electroblotted onto polyvinylidene difluoride membranes. J Biol Chem. 1987 Jul 25;262(21):10035–10038. [PubMed] [Google Scholar]
- Merril C. R., Goldman D., Sedman S. A., Ebert M. H. Ultrasensitive stain for proteins in polyacrylamide gels shows regional variation in cerebrospinal fluid proteins. Science. 1981 Mar 27;211(4489):1437–1438. doi: 10.1126/science.6162199. [DOI] [PubMed] [Google Scholar]
- Morrice L. M., McLean M. W., Long W. F., Williamson F. B. Beta-agarases I and II from Pseudomonas atlantica. Substrate specificities. Eur J Biochem. 1983 Dec 1;137(1-2):149–154. doi: 10.1111/j.1432-1033.1983.tb07808.x. [DOI] [PubMed] [Google Scholar]
- Morrice L. M., McLean M. W., Williamson F. B., Long W. F. beta-agarases I and II from Pseudomonas atlantica. Purifications and some properties. Eur J Biochem. 1983 Oct 3;135(3):553–558. doi: 10.1111/j.1432-1033.1983.tb07688.x. [DOI] [PubMed] [Google Scholar]
- Potin P., Richard C., Rochas C., Kloareg B. Purification and characterization of the alpha-agarase from Alteromonas agarlyticus (Cataldi) comb. nov., strain GJ1B. Eur J Biochem. 1993 Jun 1;214(2):599–607. doi: 10.1111/j.1432-1033.1993.tb17959.x. [DOI] [PubMed] [Google Scholar]
- Sugano Y., Matsumoto T., Kodama H., Noma M. Cloning and sequencing of agaA, a unique agarase 0107 gene from a marine bacterium, Vibrio sp. strain JT0107. Appl Environ Microbiol. 1993 Nov;59(11):3750–3756. doi: 10.1128/aem.59.11.3750-3756.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugano Y., Matsumoto T., Noma M. Sequence analysis of the agaB gene encoding a new beta-agarase from Vibrio sp. strain JT0107. Biochim Biophys Acta. 1994 May 17;1218(1):105–108. doi: 10.1016/0167-4781(94)90109-0. [DOI] [PubMed] [Google Scholar]
- Sugano Y., Terada I., Arita M., Noma M., Matsumoto T. Purification and characterization of a new agarase from a marine bacterium, Vibrio sp. strain JT0107. Appl Environ Microbiol. 1993 May;59(5):1549–1554. doi: 10.1128/aem.59.5.1549-1554.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki H., Li S. C., Li Y. T. Alpha-galactosidase from Mortierella vinacea. Crystallization and properties. J Biol Chem. 1970 Feb 25;245(4):781–786. [PubMed] [Google Scholar]
- Van Der Meulen H. J., Harder W. Characterization of the neoagarotetra-ase and neoagarobiase of Cytophaga flevensis. Antonie Van Leeuwenhoek. 1976;42(1-2):81–94. doi: 10.1007/BF00399451. [DOI] [PubMed] [Google Scholar]