Abstract
In many countries, M1 strains of the human pathogenic bacterium group A Streptococcus are the most common serotype recovered from patients with invasive disease episodes. Strains of this serotype express an extracellular protein that inhibits complement [streptococcal inhibitor of complement (Sic)] and is therefore believed to be a virulence factor. Comparative sequence analysis of the 915-bp sic gene in 165 M1 organisms recovered from diverse localities and infection types identified 62 alleles. Inasmuch as multilocus enzyme electrophoresis and pulsed-field gel electrophoresis previously showed that most M1 organisms represent a distinct streptococcal clone, the extent of sic gene polymorphism was unexpected. The level of polymorphism greatly exceeds that recorded for all other genes examined in serotype M1 strains. All insertions and deletions are in frame, and virtually all nucleotide substitutions alter the amino acid sequence of the Sic protein. These molecular features indicate that structural change in Sic is mediated by natural selection. Study of 70 strains recovered from two temporally distinct epidemics of streptococcal infections in the former East Germany found little sharing of Sic variants among strains recovered in the different time periods. Taken together, the data indicate that sic is a uniquely variable gene and provide insight into a potential molecular mechanism contributing to fluctuations in streptococcal disease frequency and severity.
Strains of the human pathogenic bacterium group A Streptococcus (GAS) are important causes of invasive disease episodes globally. The resurgence of GAS infections in the United States in recent years has led to the recognition that serotype M1 strains are the organisms most frequently recovered from normally sterile sites (1–3). Strains of this serotype are also common causes of invasive diseases in other parts of the world (reviewed in ref. 3). For example, in a 2-year study of GAS infections in Ontario, Canada, Davies et al. (4) reported that M1 strains were responsible for 24% of episodes. M1 strains generally have been the most frequently recorded serotype in surveys of invasive infections in the United Kingdom, Norway, Sweden, Finland, and elsewhere in Europe (3). They also have caused periodic peaks of invasive disease activity in New Zealand (5) and have been important causes of morbidity and mortality in Australia and Japan (6, 7). In addition to their propensity for causing invasive episodes, M1 strains also are commonly cultured from patients with pharyngitis (8) and were a prominent cause of acute rheumatic fever cases in the United States in the 1980s (9). The factors responsible for the predominance of M1 strains in human infections are unknown.
M1 organisms have been the subject of numerous molecular epidemiological investigations (3). Several techniques have been used to assess genetic variation among M1 strains, including pulsed-field gel electrophoresis (PFGE), multilocus enzyme electrophoresis, RNA gene polymorphism typing (ribotyping), random amplified polymorphic DNA analysis, and sequencing of the genes encoding streptokinase, C5a peptidase, M protein, and streptococcal pyrogenic exotoxin B (3). The common theme that has emerged from these studies is that most M1 strains cannot be subdivided by the methods used thus far (3, 10), an observation indicating a clonal relationship among the organisms.
In an analysis of molecular diversity among 17 M1 GAS strains recovered from Mexican children with pharyngitis (11), we recently identified seven alleles of the sic gene encoding an extracellular protein [streptococcal inhibitor of complement (Sic)] that inhibits human complement (12). The Sic protein is incorporated into the complement membrane–attack complex (C5b–C9) (13) and inhibits target cell lysis. The high level of polymorphism in sic was unexpected, given that other methods of molecular analysis had failed to identify substantial variation among M1 strains (10). Moreover, variation in sic sequence was detected among M1 strains that could not be subdivided on the basis of sequence analysis of genes encoding M protein, pyrogenic exotoxins A, B, and C, streptokinase, and hyaluronidase (10, 14–20).
We thought it important to study further sic allelic variation among M1 strains isolated from diverse localities and infection types. The analysis was conducted in the context of a genetic framework previously generated by multilocus enzyme electrophoresis, PFGE, and DNA sequencing of virulence genes (2, 10). We report here that the sic gene is hypervariable among M1 GAS strains recovered from intercontinental sources. Molecular variation is produced by in-frame insertion and deletion events and by nucleotide substitutions concentrated in specific regions of the gene. Virtually all nucleotide changes result in amino acid substitutions, which is indicative of natural selection mediating structural change in the Sic protein.
MATERIALS AND METHODS
Bacterial Strains.
A sample of 165 GAS strains expressing serotype M1 protein, and recovered from patients with a variety of clinical diseases, was studied. The strains were recovered from patients in 15 countries on four continents. Some of the strains were characterized previously for multilocus enzyme electrophoretic type, presence of streptococcal pyrogenic exotoxin A (speA) and speC genes, PFGE pattern, and allele of the gene (emm1) encoding M1 protein (10, 14–20).
DNA Sequence Analysis of sic.
The DNA sequencing procedures used have been described (11). The sequence data were assembled and edited with editseq, align, and seqman programs (DNAstar, Madison, WI), and the sequences were compared with a published sic sequence (12).
Variation After Laboratory Passage.
To test the hypothesis that sic variation was accumulating in the course of growth in the laboratory, we passaged three strains (MGAS 2126, MGAS 4861, and MGAS 4872) three times each on blood agar plates and then characterized the sic gene in the resulting derivative strains. A single colony was picked at each passage, streaked to a new blood agar plate, and grown overnight at 37°C. The sic sequences in the parental and laboratory passaged organisms then were determined.
Paired Isolates Cultured Sequentially from Children with Uncomplicated Pharyngitis.
The sic gene from five pairs of M1 strains cultured sequentially from children with uncomplicated pharyngitis were analyzed. These children had been enrolled in a study designed to compare the efficacy of amoxicillin and ceftibutin in treating children with uncomplicated pharyngitis. The subjects were cultured on enrollment on day 1 and again at day 10 or 17 if they were suspected to have failed therapy based on persistence of clinical disease. The children were epidemiologically unassociated.
Isolates Recovered from Epidemiologically Associated Patients.
The sic gene was analyzed in five isolates recovered from five patients in two case clusters. A strain cultured from the blood of a 4-year-old child who died of invasive GAS disease and an organism recovered 4 days later from the throat of the mother of this child were examined. The mother had performed cardiopulmonary resuscitation on the child after finding him unconscious. The other three organisms were cultured from patients who were living in the same nursing home and had experienced invasive disease episodes within 6 days of one another. Two organisms were cultured from blood and one was grown from a necrotic lesion on the arm.
RESULTS
Overall sic Variation.
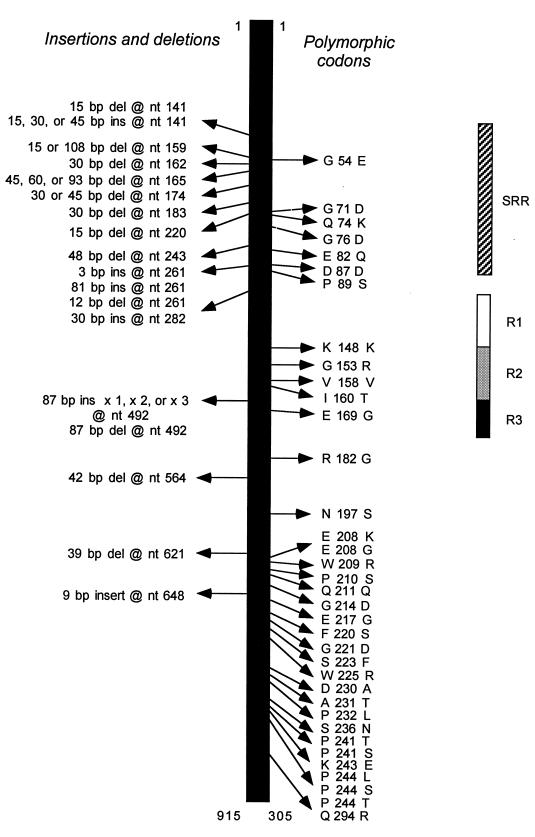
In the sample of 165 GAS strains, 62 distinct sic alleles were identified that differed by at least one nucleotide from the reference allele deposited in the GenBank database under accession no. X92968 (12). There were 36 nucleotide substitutions in 32 codons. Most of the single nucleotide changes were located in three specific parts of the gene: codons 71–89, 148–169, and 208–244 (Fig. 1). With only four exceptions, all 36 nucleotide substitutions resulted in amino acid replacements in the inferred Sic protein. Three codons had multiple mutations, all of which resulted in structural changes in Sic; compared with the sic reference sequence, codons 208 and 241 each had two variants (E → K or G; P → T or S, respectively), and codon 244 had three variants (P → L, S, or T)(Fig. 1).
Figure 1.
Summary of variation in the sic gene and Sic protein among 165 strains of serotype M1 group A Streptococcus. (Left) Positions of polymorphic nucleotides and amino acids. The single-letter amino acid abbreviations are used. (Right) Domains of Sic located in the proximal half of the protein. SRR, short repeat region; R1–R3, repeats R1, R2, and R3, respectively.
The analysis also identified 26 insertions or deletions involving from 3 to 261 bp. Most of these changes (19 of 26, 73%) were located in the first third of the gene, designated as the aminoterminal short repeat region (Fig. 1) (12). The R1-R3 repeat region also varied in length, primarily as a result of the presence of an additional 1, 2, or 3 copies of the 87-bp sequence encoding the R1 or R2 motif. In one strain, the R2 region had been deleted. All insertions and deletions were in-frame.
sic Variation and PFGE Types.
In a previous analysis (10) of genetic variation among M1 strains, chromosomal relationships among many of the organisms included in the present study were estimated by multilocus enzyme electrophoresis and PFGE profiling. That study found that two subclones [denoted ET (electrophoretic type)1/PFGE type 1a and ET 1/PFGE type 1k] commonly were recorded among organisms expressing M1 protein. Strains of ET 1/PFGE type 1a usually had the emm1.0 allele and the gene encoding pyrogenic exotoxin A (speA), whereas most ET1/PFGE type 1k organisms had allele emm1.3 but lacked speA. Organisms assigned to each subclone could not be subdivided further on the basis of sequence analysis of the genes encoding pyrogenic exotoxin B (speB) and streptokinse (ska).
Comparison of these data with the sic sequences identified in the present study revealed a substantial level of variation among strains assigned to each subclone. For example, 37 different sic alleles were found in ET1/PFGE type 1a strains; similarly, 10 sic alleles were identified among the 12 organisms of the ET1/PFGE type 1k.
Lack of Effect of Laboratory Passage on sic Variation.
Because of the unusually high degree of allelic variation in sic, we were concerned about the formal, albeit unlikely, possibility that polymorphisms might be accumulating rapidly in the course of laboratory passage. However, analysis of the sic gene in three strains passaged three times each on solid media showed no changes in sequences. In addition, we studied two strains identified as SF130 and sent to us by separate investigators. Strain SF130 originally had been cultured from a patient with scarlet fever early in this century (21), and the two derivative strains had experienced very different passage histories over the intervening decades. Nevertheless, their sic sequences (sic1.07) were identical, although distinct from those of all other strains studied.
Lack of sic Variation in Sequential Isolates Cultured from Diseased Patients.
We next determined whether sic variation occurs in vivo in the context of humans with GAS diseases. First, the gene was studied in five pairs of organisms recovered sequentially from children with uncomplicated pharyngitis. The bacteria were cultured from the pharynx on day 1 of antibiotic therapy and again on day 10 or day 17 after antibiotic therapy began. In each case, the sic alleles of the paired isolates were identical. To investigate further the possibility that sic variation occurs rapidly in vivo, we sequenced the gene in strains recovered from epidemiologically linked patients with GAS disease. The genes in organisms cultured from a child with invasive disease and his mother were identical, as were the genes of three GAS strains recovered from associated patients living in a nursing home. In sum, the results showed that sic variation did not accumulate in the short time frames represented by these epidemiological situations.
sic Variation and Temporal Changes in Disease Frequency.
To determine whether there is a relationship between Sic variation and temporal change in disease frequency, we studied strains recovered from patients in the former East Germany, a geographic region where the epidemiology of disease caused by M1 strains has been studied extensively since the 1960s (22). Beginning late in 1975, M1 strains increased in frequency from ≈3 to 25% of all strains recovered from diseased patients. M1 strains continued to be recovered at relatively high frequency through 1978, at which time they decreased in frequency to ≈5% of isolates. M1 strains then again increased in frequency in early 1981 and peaked at 40–45% of isolates in 1982–1983. Previous analysis of genes encoding pyrogenic exotoxin A, exotoxin B, streptokinase, and M protein failed to detect allelic variation among organisms recovered within these two time frames (14); nor could strains be differentiated by multilocus enzyme electrophoresis or PFGE (10).
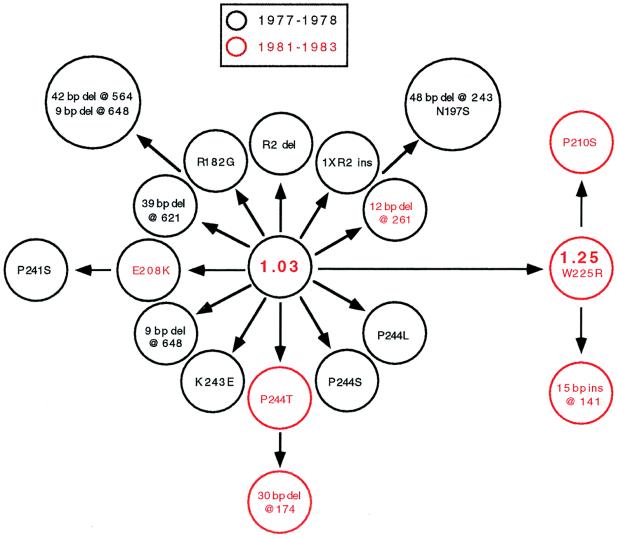
We tested the possibility that allele frequency variation between populations of strains was associated with the two outbreaks in the former East Germany. First, sic was sequenced in 37 M1T1 strains cultured from patients residing in 19 communities in 1977 and 1978. Unexpectedly, 15 different sic alleles were identified. The most common allele was sic1.03, and this variant occurred in 20 of the 37 strains (54%). We then characterized sic in 33 M1T1 strains recovered from patients living in 18 communities in 1981–1983. To minimize sampling bias, we selected organisms cultured from patients residing in the same towns as those studied from the 1977–1978 outbreak. A total of 13 alleles was identified among the 1981–1983 sample, but only nine of the strains (27%) had the sic1.03 allele that commonly was represented among organisms recovered in 1977–1978, and 8 organisms (24%) had an allele (sic1.25) that was not represented in the earlier sample. The sic1.25 allele differs from the sic 1.03 allele by a single nucleotide change that produces an amino acid substitution at Sic position 225 (TGG → CGG, Trp → Arg). All communities from which strains with sic1.25 were drawn also were represented in the 1977–1978 sample, which rules out the possibility that the failure to identify sic1.25 in the earlier period was a result of sampling strains recovered in different communities. In addition, with only two other exceptions (sic1.26 and sic1.40), there was no sharing of alleles among M1 strains cultured from patients in the two distinct disease-peak periods. Moreover, only 3 of the total of 25 sic alleles represented in the 70 strains from the former East Germany were found in the other 95 organisms in the sample.
Inspection of the sic sequence data and inferred Sic amino acid sequences revealed that most alleles readily could be linked to the commonly occurring sic1.03 and sic1.25 variants by one or two structural changes (Fig. 2). For example, seven alleles differed from sic1.03 by single nucleotide changes, each of which results in an amino acid replacement. Taken together, the occurrence of a large number of Sic variants differing from one another by structural changes that involve only one or two mutational steps and our failure to identify these Sic variants in areas other than the former East Germany suggest that most of them arose in the course of the distinct waves of M1 disease.
Figure 2.
Hypothetical scheme for the evolution of Sic variants associated with temporal variation in disease frequency in the former East Germany. Strains recovered in two time periods (1977–1978 and 1981–1983) were studied. The analysis identified 18 Sic variants that could be linked to Sic1.03 by one or two structural changes. M1 strains expressing Sic1.03 protein caused 54% of disease episodes in 1977–1978. The Sic1.03 protein is hypothesized to be ancestral because it was abundant. Strains expressing Sic1.25 did not occur in the 1977–1978 sample but were recovered from 24% of patients in the 1981–1983 sample. There was little sharing of Sic variants between the two time periods. Single-letter amino acid abbreviations are used, and the variant amino acid residues are designated. The locations of the insertion and deletion events are indicated by nucleotide position. Sic variants recovered in both time periods are denoted by red text within a black circle. Two of the circles are larger than the other 16 to accommodate text. del, deletion; ins, insertion.
DISCUSSION
With 62 alleles represented among 165 strains, the sic gene is extremely and uniquely polymorphic among the 12 genes that have been sequenced comparatively in clonally related M1 strains (Table 1). Notably, the level of variation far exceeds that of the aminoterminal hypervariable region of the M protein, the part of the molecule against which serotype-specific antibody is made (10, 23, 24) (unpublished data).
Table 1.
Levels of sequence variation in genes in ET1/M1 group A Streptococcus
| Gene | Size, bp* | Product | Strains†, n | Polymorphic sites |
|---|---|---|---|---|
| speA | 708 | Pyrogenic exotoxin A | 345 | 1 |
| speB | 1,484 | Pyrogenic exotoxin B | 26 | 0 |
| speC | 702 | Pyrogenic exotoxin C | 44 | 1 |
| ska | 513 | Streptokinase | 18 | 0 |
| scp | 416 | C5a peptidase | 14 | 0 |
| pcp | 650 | Pyrrolidine carboxyl peptidase | 8 | 0 |
| hyl | 1,116 | Hyaluronidase | 4 | 0 |
| ssa | 783 | Streptococcal superantigen | 14 | 0 |
| rrs | 1,464 | 16S rRNA | 6 | 0 |
| emm | 300 | M protein, variable region | 214 | 8 |
| sic | 915 | Complement inhibitory protein | 165 | 62 |
Geographic Variation in Frequency of Occurrence of sic Alleles.
We found marked geographic variation in the frequency of sic alleles. Strains expressing the same sic allele rarely were recovered from patients living on different continents or in different regions of the same continent. Organisms with allele sic1.01 were the only notable exceptions to this general observation; sic1.01 occurred in highest frequency in our sample and was present in strains cultured from patients in five states. The concept of geographic variation in sic allele frequency is exemplified by the strains from the former East Germany. Among the 70 strains recovered from the two time periods sampled, 25 alleles were identified, but only three of them were found in strains from other countries. Inasmuch as GAS cannot disseminate instantaneously over large areas or intercontinentally, geographic variation in sic allele frequency is the expected condition if sic alleles are being generated in the course of host–pathogen interactions.
Sic Variation and PFGE Patterns.
It is now common practice to analyze the PFGE patterns of bacterial strains for epidemiological purposes and for inferring relationships among organisms, including GAS (25, 26). Attempts have been made to develop interpretive criteria, but there is controversy regarding the rationale and performance of this technique for assessing strain relationships (27). One frequently cited concern is that relatively few nucleotides are directly indexed for variation. Previously, we reported that 16 PFGE patterns could be distinguished among 126 M1 strains obtained from intercontinental sources (10). However, two related PFGE patterns (designated 1a and 1k) accounted for 83 (66%) and 17 (13%), respectively, of the 126 organisms studied. Although not all strains analyzed in the present study had been characterized by PFGE, those organisms for which data were available included a total of 37 sic alleles among isolates of PFGE type 1a and 10 alleles among PFGE type 1k organisms. Hence, the data indicate that considerable sic variation can exist among isolates with the same PFGE pattern.
Implications for Understanding Temporal Variation in Disease Frequency.
In recent years, strains of GAS expressing M1 protein repeatedly have been recovered from episodes of invasive disease in many countries (3). The most commonly implicated M1 organisms are ET1/PFGE type 1a and express exotoxin A (3, 10). Investigators in several countries have reported that strains expressing M1 protein can increase rapidly in frequency of recovery from disease episodes (3, 5, 22). But despite years of study, the molecular basis for this phenomenon remains unknown. Nor is there any insight into the mechanisms responsible for the abundance of M1 strains in human infections.
Several lines of evidence suggest that Sic variation may participate in temporal variation in disease frequency. First, we have discovered that there is a large pool of genetic subclones marked by the ET 1/M1/speA-positive character combination. This heterogenous array of sublines provides an abundant, geographically dispersed pool of organisms from which a more fit cell line can be derived. Second, analysis of the M1 strains recovered from the two distinct scarlet fever epidemics in the former East Germany demonstrated that each disease peak was characterized by GAS strains expressing predominantly different sic alleles. Organisms cultured from these two peaks of disease could not otherwise be differentiated by biochemical or genetic methods, including the sequencing of genes encoding proven or presumed virulence factors (10, 14–20). Important to note, most isolates from the two disease peaks had the same sequence in the hypervariable region of the emm gene encoding M protein (unpublished data), a molecule known to be under intense selective pressure because of the host immune response (23, 24).
sic Variation and Interaction with Host and Pathogen Proteins.
The most notable aspects of variation in the sic gene are that all insertions and deletions are in-frame and that almost all nucleotide substitutions result in amino acid substitutions. This situation is unusual because, for bacterial genes in general, synonymous (silent) nucleotide changes outnumber nonsynonymous changes by a factor of 10 to 20 (28). The magnitude of this factor varies and depends on evolutionary forces and functional constraints. An unusually high level of polymorphism is characteristic of bacterial genes encoding exported molecules that directly interact with the environment, such as certain virulence factors (29, 30). However, even among strongly polymorphic genes encoding virulence factors, there is no precedent for the high levels of sic allelic variation identified among strains assigned to the same multilocus enzyme electrophoretic type, combined with the finding that essentially all gene variation results in structural changes in the protein product.
For a selectively neutral gene, nucleotide substitutions are expected to be distributed randomly across the three codon positions; that is, roughly one-third of the changes should occur at each position, in which case ≈70% of them would be nonsynonymous. For sic, we tested the hypothesis of selective neutrality by comparing the expected and observed numbers of nucleotide changes occurring in the first, second, and third codon positions. Most (32 of 36) substitutions occurred in the first or second position, a result permitting us to reject the null hypothesis of neutrality (χ2(2) = 8.12; P < 0.025). Since this study was completed, sequence analysis of sic from 218 M1 isolates cultured from patients in Ontario, Canada and Japan has identified 13 new polymorphic nucleotides, all of which produce amino acid substitutions (unpublished data).
There are parallels between sic variation and that identified in major histocompatibility complex genes that encode cell surface glycoproteins that function to present peptides to cytotoxic T cells and are among the more highly polymorphic genes known (31). As we have discovered for Sic, there is evidence that natural selection is responsible for a high degree of polymorphism in the functionally important peptide-binding region (32, 33). In regard to proteins expressed by pathogenic microbes, evidence has been presented that positive selection is acting on immunogenic regions of the cell surface-associated circumsporozoite protein made by malaria parasites and the merozoite surface antigen-1 of Plasmodium falciparum (34) and on those regions of several proteins made by HIV-1 (35). We also note that 18 of the 32 amino acid substitutions in Sic would result in charge differences and that 10 of the replacements change a polar to a nonpolar amino acid or vice versa. This pattern of amino acid replacements is also consistent with the idea that positive selection is favoring diversity at the protein level in Sic (34, 36).
Inasmuch as natural selection of amino acid substitutions is the most likely hypothesis to explain the characteristics of allelic diversity in sic, a question arises regarding when selection is operative during host–pathogen interaction. Sic has a leader sequence associated with secreted proteins; lacks an amino acid motif (LPXTG) characteristic of many Gram-positive, cell wall-anchored proteins (37); and can be purified from growth medium (12). Hence, one can speculate that humans mount an antibody response to Sic, a process that might be expected to enhance variation by selecting for escape mutants. However, we favor the hypothesis that Sic variation is promoted by the ability of the protein to inhibit complement. Akesson et al. (12) reported that the Sic protein is incorporated into the complement membrane-attack complex (C5b-C9) (13), thereby suggesting the site at which Sic inhibits target cell lysis. One implication from the data is that Sic variants with an elevated ability to block complement function may have enhanced survival in vivo. Inasmuch as the individual proteins of the complement system are themselves polymorphic (31, 38–41), it is also possible that nonrandom interactions occur among variants of Sic and target complement proteins. Differences in these types of protein–protein interactions could significantly alter the outcome of the interplay between host and pathogen.
Recently, there has been interest in the idea that some emerging bacterial pathogens may have unusually high mutation frequencies (42, 43). LeClerc et al. (42) reported that >1% of pathogenic Escherichia coli and Salmonella enterica isolates are hypermutable because of defects in methyl-directed mismatch DNA repair. Similarly, Sniegowski et al. (43) found in chemostat experiments conducted with E. coli that progeny with a stable mutator phenotype could arise from the ancestral strain. These observations raise the possibility that hypervariability in Sic is caused, in part, by an elevated mutation frequency. However, a simple hypermutation hypothesis alone is untenable for two reasons. First, none of the other loci we studied for sequence variation had levels of allelic variation approaching that of the sic gene, and, indeed, most showed no variation. Second, hypermutation in the absence of selection would not account for the observed preponderance of protein-altering mutations relative to silent nucleotide changes.
Concluding Comment.
In summary, the present study makes several contributions to our understanding of the biology of GAS. First, the data demonstrate that sic is by far the most polymorphic gene yet identified among strains expressing the M1 protein, a serotype responsible for a large percentage of invasive episodes in North America and Europe. Second, almost all nucleotide variation results in changes in the amino acid sequence of the Sic protein, which we interpret as reflecting the action of natural selection in adapting the Sic protein to variable host factors. Third, an analysis of M1 strains causing streptococcal epidemics in the former East Germany shows that Sic variation is associated with temporal shifts in disease frequency. This observation suggests that Sic variation contributes to new epidemics of disease caused by serotype M1 strains, although more work is needed to investigate this hypothesis. Inasmuch as we have essentially no molecular insight into the processes responsible for rapid changes in M1 disease frequency and character, it is important to elucidate the mechanisms responsible for sic polymorphism and the functional consequences of Sic variation.
Acknowledgments
We thank our many colleagues who contributed bacterial strains, D. Meyer for secretarial assistance, and M. Majesky for critical comments. We are indebted to P. McInnes for ongoing support and to Professor R. K. Selander for editorial assistance. This work is dedicated to P. D. Bennett and R. K. Selander on the occasions of retirement and seventieth birthday, respectively. This study was supported by Public Health Service Grant AI-33119, Fogarty International Center Grant TW-37004-03S1, and North Atlantic Treaty Organization Collaborative Research Grant 920248. J.M.M. is an Established Investigator of the American Heart Association.
ABBREVIATIONS
- GAS
group A Streptococcus
- PFGE
pulsed-field gel electrophoresis
- ET
electrophoretic type
- SPE
streptococcal pyrogenic exotoxin
- Sic
streptococcal inhibitor of complement
References
- 1.Schwartz B, Facklam R R, Breiman R F. Lancet. 1990;336:1167–1171. doi: 10.1016/0140-6736(90)92777-f. [DOI] [PubMed] [Google Scholar]
- 2.Musser J M, Hauser A R, Kim M H, Schlievert P M, Nelson K, Selander R K. Proc Natl Acad Sci USA. 1991;88:2668–2672. doi: 10.1073/pnas.88.7.2668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Musser J M, Krause R M. In: Emerging Infections. Krause R M, Fauci A, editors. San Diego: Academic; 1998. pp. 185–218. [Google Scholar]
- 4.Davies H D, McGeer A, Schwartz B, Green K, Cann D, Simor A E, Low D E The Ontario Group A Streptococcal Study Group. N Engl J Med. 1996;335:547–554. doi: 10.1056/NEJM199608223350803. [DOI] [PubMed] [Google Scholar]
- 5.Martin D R, Single L A. J Infect Dis. 1993;167:1112–1117. doi: 10.1093/infdis/167.5.1112. [DOI] [PubMed] [Google Scholar]
- 6.Carapetis J R, Robins-Browne R, Martin D, Shelby-James T, Hogg G. Clin Infect Dis. 1995;21:1220–1227. doi: 10.1093/clinids/21.5.1220. [DOI] [PubMed] [Google Scholar]
- 7.Nakashima K, Ichiyama S, Iinuma Y, Hasegawa Y, Ohta M, Ooe K, Shimizu Y, Igarashi H, Murai T, Shimokata K. Clin Infect Dis. 1997;25:260–266. doi: 10.1086/514543. [DOI] [PubMed] [Google Scholar]
- 8.Johnson D R, Stevens D L, Kaplan E L. J Infect Dis. 1992;166:374–382. doi: 10.1093/infdis/166.2.374. [DOI] [PubMed] [Google Scholar]
- 9.Kaplan E L, Johnson D R, Cleary P P. J Infect Dis. 1989;159:101–103. doi: 10.1093/infdis/159.1.101. [DOI] [PubMed] [Google Scholar]
- 10.Musser J M, Kapur V, Szeto J, Pan X, Swanson D, Martin D R. Infect Immun. 1995;63:994–1003. doi: 10.1128/iai.63.3.994-1003.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Perea Mejia L M, Stockbauer K E, Pan X, Cravioto A, Musser J M. J Clin Microbiol. 1997;35:3220–3224. doi: 10.1128/jcm.35.12.3220-3224.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Akesson P, Sjoholm A G, Bjorck L. J Biol Chem. 1996;271:1081–1088. doi: 10.1074/jbc.271.2.1081. [DOI] [PubMed] [Google Scholar]
- 13.Muller-Eberhard H J. Annu Rev Immunol. 1986;4:503–528. doi: 10.1146/annurev.iy.04.040186.002443. [DOI] [PubMed] [Google Scholar]
- 14.Kapur V, Nelson K, Schlievert P M, Selander R K, Musser J M. Infect Immun. 1992;60:3513–3517. doi: 10.1128/iai.60.9.3513-3517.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Musser J M, Nelson K, Selander R K, Gerlach D, Huang J C, Kapur V, Kanjilal S. J Infect Dis. 1993;167:759–762. doi: 10.1093/infdis/167.3.759. [DOI] [PubMed] [Google Scholar]
- 16.Kapur V, Topouzis S, Majesky M W, Li L-L, Hamrick M R, Hamill R J, Patti J M, Musser J M. Microb Pathog. 1993;15:327–346. doi: 10.1006/mpat.1993.1083. [DOI] [PubMed] [Google Scholar]
- 17.Marciel A M, Kapur V, Musser J M. Microb Pathog. 1997;22:209–217. doi: 10.1006/mpat.1996.9999. [DOI] [PubMed] [Google Scholar]
- 18.Kapur V, Kanjilal S, Hamrick M R, Li L-L, Whittam T S, Sawyer S A, Musser J M. Mol Microbiol. 1995;16:509–519. doi: 10.1111/j.1365-2958.1995.tb02415.x. [DOI] [PubMed] [Google Scholar]
- 19.Nelson K, Schlievert P M, Selander R K, Musser J M. J Exp Med. 1991;174:1271–1274. doi: 10.1084/jem.174.5.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Musser J M, Kapur V, Kanjilal S, Shah U, Musher D M, Barg N L, Johnston K H, Schlievert P M, Henrichsen J, Gerlach D, et al. J Infect Dis. 1993;167:337–346. doi: 10.1093/infdis/167.2.337. [DOI] [PubMed] [Google Scholar]
- 21.Griffith F. J Hyg. 1926;25:385–397. doi: 10.1017/s0022172400017502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kohler W, Gerlach D, Knoll H. Zentralbl Bakteriol Hyg A. 1987;266:104–115. doi: 10.1016/s0176-6724(87)80024-x. [DOI] [PubMed] [Google Scholar]
- 23.Lancefield R C. J Immunol. 1962;89:307–313. [PubMed] [Google Scholar]
- 24.Fischetti V A. Clin Microbiol Rev. 1989;2:285–314. doi: 10.1128/cmr.2.3.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tenover F C, Arbeit R D, Goering R V, Mickelsen P A, Murray B E, Persing D H, Swaminathan B. J Clin Microbiol. 1995;33:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cockerill F R, III, MacDonald K L, Thompson R L, Roberson F, Kohner PC, Besser-Wiek J, Manahan J M, Musser J M, Schlievert P M, Talbot J, et al. J Am Med Assoc. 1997;277:38–43. [PubMed] [Google Scholar]
- 27.Petts D N, Noble W C, Howell S A. J Clin Microbiol. 1997;35:1722–1727. doi: 10.1128/jcm.35.7.1722-1727.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ochman H, Wilson A C. J Mol Evol. 1987;26:74–86. doi: 10.1007/BF02111283. [DOI] [PubMed] [Google Scholar]
- 29.Whatmore A M, Kapur V, Sullivan D J, Musser J M, Kehoe M A. Mol Microbiol. 1994;14:619–631. doi: 10.1111/j.1365-2958.1994.tb01301.x. [DOI] [PubMed] [Google Scholar]
- 30.Li J, Ochman H, Groisman E A, Boyd E F, Solomon F, Nelson K, Selander R K. Proc Natl Acad Sci USA. 1995;92:7252–7256. doi: 10.1073/pnas.92.16.7252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Klein J, Klein D. Molecular Evolution of the Major Histocompatibility Complex. Heidelberg: Springer; 1991. [Google Scholar]
- 32.Hughes A L, Nei M. Nature (London) 1988;335:167–170. doi: 10.1038/335167a0. [DOI] [PubMed] [Google Scholar]
- 33.Hughes A L, Nei M. Proc Natl Acad Sci USA. 1989;86:958–962. doi: 10.1073/pnas.86.3.958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hughes M K, Hughes A L. Mol Biochem Parasitol. 1995;71:99–113. doi: 10.1016/0166-6851(95)00037-2. [DOI] [PubMed] [Google Scholar]
- 35.Hughes A L, Ota T, Nei M. Mol Biol Evol. 1990;7:515–524. doi: 10.1093/oxfordjournals.molbev.a040626. [DOI] [PubMed] [Google Scholar]
- 36.Seibert S A, Howell C Y, Hughes M K, Hughes A L. Mol Biol Evol. 1995;12:803–813. doi: 10.1093/oxfordjournals.molbev.a040257. [DOI] [PubMed] [Google Scholar]
- 37.Schneewind O, Model P, Fischetti V A. Cell. 1992;70:267–281. doi: 10.1016/0092-8674(92)90101-h. [DOI] [PubMed] [Google Scholar]
- 38.Nakamura S, Ooue O, Abe K. Hum Genet. 1984;66:279–281. doi: 10.1007/BF00286617. [DOI] [PubMed] [Google Scholar]
- 39.York L J, Marshall W H, Huang S N. Hum Hered. 1986;36:261–262. doi: 10.1159/000153639. [DOI] [PubMed] [Google Scholar]
- 40.Fernie B A, Finlay A, Price D, Chan E, Orren A, Joysey V C, Joysey K A, Hobart M J. Exp Clin Immunogenet. 1996;13:92–103. [PubMed] [Google Scholar]
- 41.Herrmann D, Sodetz J M, Rittner C, Schneider P M. Immunogenetics. 1989;30:291–295. doi: 10.1007/BF02421333. [DOI] [PubMed] [Google Scholar]
- 42.LeClerc J E, Li B, Payne W L, Cebula T A. Science. 1996;274:1208–1211. doi: 10.1126/science.274.5290.1208. [DOI] [PubMed] [Google Scholar]
- 43.Sniegowski P D, Gerrish P J, Lenski R E. Nature (London) 1997;387:703–705. doi: 10.1038/42701. [DOI] [PubMed] [Google Scholar]