Abstract
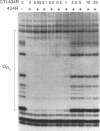
The repressor protein of bacteriophage 434 binds to DNA as a dimer of identical subunits. Its strong dimerization is mediated by the carboxyl-terminal domain. Cooperative interactions between the C-terminal domains of two repressor dimers bound at adjacent sites can stabilize protein-DNA complexes formed with low-affinity binding sites. We have constructed a plasmid, pCT1, which directs the overproduction of the carboxyl-terminal domain of 434 repressor. The protein encoded by this plasmid is called CT-1. Cells transformed with pCT1 are unable to be lysogenized by wild-type 434 phage, whereas control cells are lysogenized at an efficiency of 1 to 5%. The CT-1-mediated interference with lysogen formation presumably results from formation of heteromeric complexes between the phage-encoded repressor and the plasmid-encoded carboxyl-terminal domain fragment. These heteromers are unable to bind DNA and thereby inhibit the repressor's activity in promoting lysogen formation. Two lines of evidence support this conclusion. First, DNase I footprinting experiments show that at a 2:1 ratio of CT-1 to intact 434 repressor, purified CT-1 protein prevents the formation of complexes between 434 repressor and its OR1 binding site. Second, cross-linking experiments reveal that only a specific heterodimeric complex forms between CT-1 and intact 434 repressor. This latter observation indicates that CT-1 interferes with 434 repressor-operator complex formation by preventing dimerization and not by altering the conformation of the DNA-bound repressor dimer. Our other evidence is also consistent with this suggestion. We have used deletion analysis in an attempt to define the region which mediates the 434 repressor-CT-1 interaction. CT-1 proteins which have more than the last 14 amino acids removed are unable to interfere with 434 repressor action in vivo.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aggarwal A. K., Rodgers D. W., Drottar M., Ptashne M., Harrison S. C. Recognition of a DNA operator by the repressor of phage 434: a view at high resolution. Science. 1988 Nov 11;242(4880):899–907. doi: 10.1126/science.3187531. [DOI] [PubMed] [Google Scholar]
- Anderson J., Ptashne M., Harrison S. C. Cocrystals of the DNA-binding domain of phage 434 repressor and a synthetic phage 434 operator. Proc Natl Acad Sci U S A. 1984 Mar;81(5):1307–1311. doi: 10.1073/pnas.81.5.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckett D., Burz D. S., Ackers G. K., Sauer R. T. Isolation of lambda repressor mutants with defects in cooperative operator binding. Biochemistry. 1993 Sep 7;32(35):9073–9079. doi: 10.1021/bi00086a012. [DOI] [PubMed] [Google Scholar]
- Bell A. C., Koudelka G. B. Operator sequence context influences amino acid-base-pair interactions in 434 repressor-operator complexes. J Mol Biol. 1993 Dec 5;234(3):542–553. doi: 10.1006/jmbi.1993.1610. [DOI] [PubMed] [Google Scholar]
- Benson N., Adams C., Youderian P. Genetic selection for mutations that impair the co-operative binding of lambda repressor. Mol Microbiol. 1994 Feb;11(3):567–579. doi: 10.1111/j.1365-2958.1994.tb00337.x. [DOI] [PubMed] [Google Scholar]
- Coulondre C., Miller J. H. Genetic studies of the lac repressor. III. Additional correlation of mutational sites with specific amino acid residues. J Mol Biol. 1977 Dec 15;117(3):525–567. doi: 10.1016/0022-2836(77)90056-0. [DOI] [PubMed] [Google Scholar]
- De Anda J., Poteete A. R., Sauer R. T. P22 c2 repressor. Domain structure and function. J Biol Chem. 1983 Sep 10;258(17):10536–10542. [PubMed] [Google Scholar]
- Finger L. R., Richardson J. P. Stabilization of the hexameric form of Escherichia coli protein rho under ATP hydrolysis conditions. J Mol Biol. 1982 Mar 25;156(1):203–219. doi: 10.1016/0022-2836(82)90467-3. [DOI] [PubMed] [Google Scholar]
- Hochschild A., Ptashne M. Interaction at a distance between lambda repressors disrupts gene activation. Nature. 1988 Nov 24;336(6197):353–357. doi: 10.1038/336353a0. [DOI] [PubMed] [Google Scholar]
- Johnson A. D., Meyer B. J., Ptashne M. Interactions between DNA-bound repressors govern regulation by the lambda phage repressor. Proc Natl Acad Sci U S A. 1979 Oct;76(10):5061–5065. doi: 10.1073/pnas.76.10.5061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koudelka G. B., Carlson P. DNA twisting and the effects of non-contacted bases on affinity of 434 operator for 434 repressor. Nature. 1992 Jan 2;355(6355):89–91. doi: 10.1038/355089a0. [DOI] [PubMed] [Google Scholar]
- Koudelka G. B., Harrison S. C., Ptashne M. Effect of non-contacted bases on the affinity of 434 operator for 434 repressor and Cro. 1987 Apr 30-May 6Nature. 326(6116):886–888. doi: 10.1038/326886a0. [DOI] [PubMed] [Google Scholar]
- Lim W. A., Farruggio D. C., Sauer R. T. Structural and energetic consequences of disruptive mutations in a protein core. Biochemistry. 1992 May 5;31(17):4324–4333. doi: 10.1021/bi00132a025. [DOI] [PubMed] [Google Scholar]
- Lim W. A., Sauer R. T. The role of internal packing interactions in determining the structure and stability of a protein. J Mol Biol. 1991 May 20;219(2):359–376. doi: 10.1016/0022-2836(91)90570-v. [DOI] [PubMed] [Google Scholar]
- Valenzuela D., Ptashne M. P22 repressor mutants deficient in co-operative binding and DNA loop formation. EMBO J. 1989 Dec 20;8(13):4345–4350. doi: 10.1002/j.1460-2075.1989.tb08621.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wharton R. P., Ptashne M. Changing the binding specificity of a repressor by redesigning an alpha-helix. Nature. 1985 Aug 15;316(6029):601–605. doi: 10.1038/316601a0. [DOI] [PubMed] [Google Scholar]