Abstract
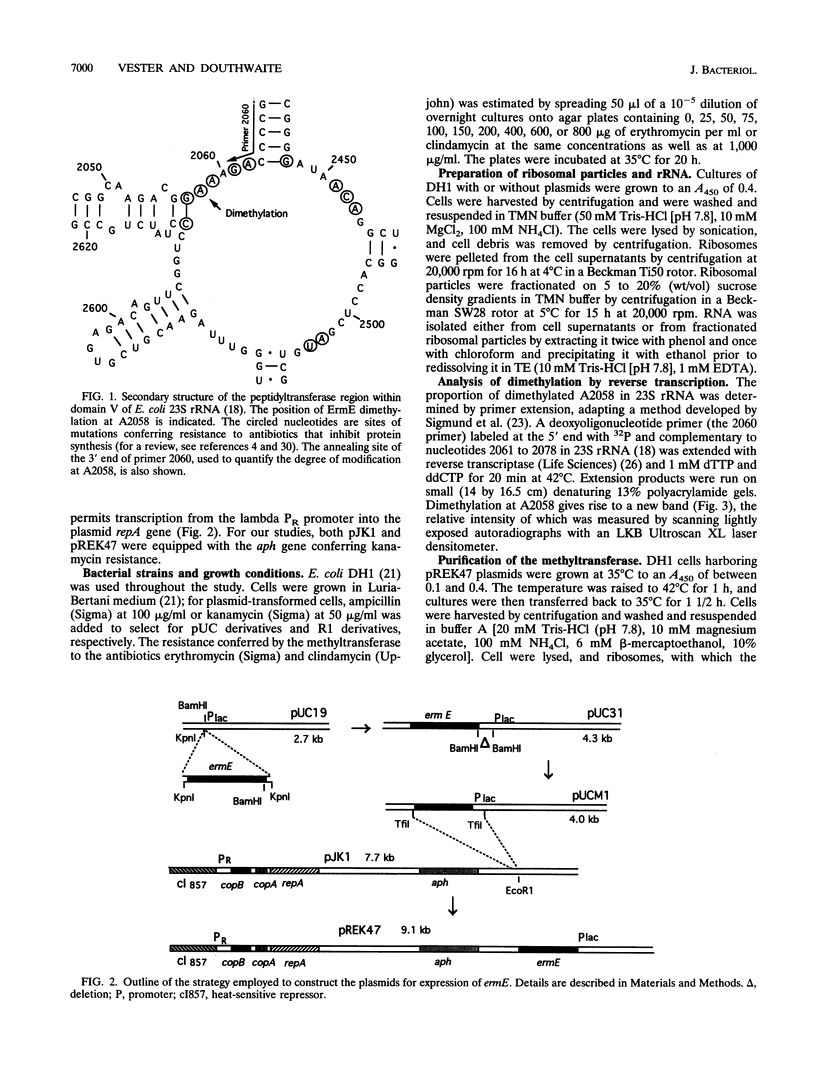
The ErmE methyltransferase from the erythromycin-producing actinomycete Saccharopolyspora erythraea dimethylates the N-6 position of adenine 2058 in domain V of 23S rRNA. This modification confers resistance to erythromycin and to other macrolide, lincosamide, and streptogramin B antibiotics. We investigated what structural elements in 23S rRNA are required for specific recognition by the ErmE methyltransferase. The ermE gene was cloned into R1 plasmid derivatives, providing a means of inducible expression in Escherichia coli. Expression of the methyltransferase in vivo confers resistance to erythromycin and clindamycin. The degree of resistance corresponds to the level of ermE expression. In turn, ermE expression also correlates with the proportion of 23S rRNA molecules that are dimethylated at adenine 2058. The methyltransferase was isolated in an active, concentrated form from E. coli, and the enzyme efficiently modifies 23S rRNA in vitro. Removal of most of the 23S rRNA structure, so that only domain V (nucleotides 2000 to 2624) remains, does not affect the efficiency of modification by the methyltransferase. In addition, modification still occurs after the rRNA tertiary structure has been disrupted by removal of magnesium ions. We conclude that the main features that are specifically recognized by the ErmE methyltransferase are displayed within the primary and secondary structures of 23S rRNA domain V.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Douthwaite S., Aagaard C. Erythromycin binding is reduced in ribosomes with conformational alterations in the 23 S rRNA peptidyl transferase loop. J Mol Biol. 1993 Aug 5;232(3):725–731. doi: 10.1006/jmbi.1993.1426. [DOI] [PubMed] [Google Scholar]
- Douthwaite S. Functional interactions within 23S rRNA involving the peptidyltransferase center. J Bacteriol. 1992 Feb;174(4):1333–1338. doi: 10.1128/jb.174.4.1333-1338.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubnau D. Translational attenuation: the regulation of bacterial resistance to the macrolide-lincosamide-streptogramin B antibiotics. CRC Crit Rev Biochem. 1984;16(2):103–132. doi: 10.3109/10409238409102300. [DOI] [PubMed] [Google Scholar]
- Gutell R. R., Gray M. W., Schnare M. N. A compilation of large subunit (23S and 23S-like) ribosomal RNA structures: 1993. Nucleic Acids Res. 1993 Jul 1;21(13):3055–3074. doi: 10.1093/nar/21.13.3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutell R. R., Woese C. R. Higher order structural elements in ribosomal RNAs: pseudo-knots and the use of noncanonical pairs. Proc Natl Acad Sci U S A. 1990 Jan;87(2):663–667. doi: 10.1073/pnas.87.2.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz L., Brown D., Boris K., Tuan J. Expression of the macrolide-lincosamide-streptogramin-B-resistance methylase gene, ermE, from Streptomyces erythraeus in Escherichia coli results in N6-monomethylation and N6,N6-dimethylation of ribosomal RNA. Gene. 1987;55(2-3):319–325. doi: 10.1016/0378-1119(87)90291-5. [DOI] [PubMed] [Google Scholar]
- Larsen J. E., Gerdes K., Light J., Molin S. Low-copy-number plasmid-cloning vectors amplifiable by derepression of an inserted foreign promoter. Gene. 1984 Apr;28(1):45–54. doi: 10.1016/0378-1119(84)90086-6. [DOI] [PubMed] [Google Scholar]
- Larsen N. Higher order interactions in 23s rRNA. Proc Natl Acad Sci U S A. 1992 Jun 1;89(11):5044–5048. doi: 10.1073/pnas.89.11.5044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen N., Olsen G. J., Maidak B. L., McCaughey M. J., Overbeek R., Macke T. J., Marsh T. L., Woese C. R. The ribosomal database project. Nucleic Acids Res. 1993 Jul 1;21(13):3021–3023. doi: 10.1093/nar/21.13.3021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leffers H., Egebjerg J., Andersen A., Christensen T., Garrett R. A. Domain VI of Escherichia coli 23 S ribosomal RNA. Structure, assembly and function. J Mol Biol. 1988 Dec 5;204(3):507–522. doi: 10.1016/0022-2836(88)90351-8. [DOI] [PubMed] [Google Scholar]
- Moazed D., Noller H. F. Chloramphenicol, erythromycin, carbomycin and vernamycin B protect overlapping sites in the peptidyl transferase region of 23S ribosomal RNA. Biochimie. 1987 Aug;69(8):879–884. doi: 10.1016/0300-9084(87)90215-x. [DOI] [PubMed] [Google Scholar]
- Noller H. F. Structure of ribosomal RNA. Annu Rev Biochem. 1984;53:119–162. doi: 10.1146/annurev.bi.53.070184.001003. [DOI] [PubMed] [Google Scholar]
- Peattie D. A., Gilbert W. Chemical probes for higher-order structure in RNA. Proc Natl Acad Sci U S A. 1980 Aug;77(8):4679–4682. doi: 10.1073/pnas.77.8.4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosendahl G., Douthwaite S. Ribosomal proteins L11 and L10.(L12)4 and the antibiotic thiostrepton interact with overlapping regions of the 23 S rRNA backbone in the ribosomal GTPase centre. J Mol Biol. 1993 Dec 20;234(4):1013–1020. doi: 10.1006/jmbi.1993.1655. [DOI] [PubMed] [Google Scholar]
- Sigmund C. D., Ettayebi M., Borden A., Morgan E. A. Antibiotic resistance mutations in ribosomal RNA genes of Escherichia coli. Methods Enzymol. 1988;164:673–690. doi: 10.1016/s0076-6879(88)64077-8. [DOI] [PubMed] [Google Scholar]
- Skinner R., Cundliffe E., Schmidt F. J. Site of action of a ribosomal RNA methylase responsible for resistance to erythromycin and other antibiotics. J Biol Chem. 1983 Oct 25;258(20):12702–12706. [PubMed] [Google Scholar]
- Stern S., Moazed D., Noller H. F. Structural analysis of RNA using chemical and enzymatic probing monitored by primer extension. Methods Enzymol. 1988;164:481–489. doi: 10.1016/s0076-6879(88)64064-x. [DOI] [PubMed] [Google Scholar]
- Su S. L., Dubnau D. Binding of Bacillus subtilis ermC' methyltransferase to 23S rRNA. Biochemistry. 1990 Jun 26;29(25):6033–6042. doi: 10.1021/bi00477a022. [DOI] [PubMed] [Google Scholar]
- Uchiyama H., Weisblum B. N-Methyl transferase of Streptomyces erythraeus that confers resistance to the macrolide-lincosamide-streptogramin B antibiotics: amino acid sequence and its homology to cognate R-factor enzymes from pathogenic bacilli and cocci. Gene. 1985;38(1-3):103–110. doi: 10.1016/0378-1119(85)90208-2. [DOI] [PubMed] [Google Scholar]
- Vester B., Garrett R. A. The importance of highly conserved nucleotides in the binding region of chloramphenicol at the peptidyl transfer centre of Escherichia coli 23S ribosomal RNA. EMBO J. 1988 Nov;7(11):3577–3587. doi: 10.1002/j.1460-2075.1988.tb03235.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisblum B. Inducible resistance to macrolides, lincosamides and streptogramin type B antibiotics: the resistance phenotype, its biological diversity, and structural elements that regulate expression--a review. J Antimicrob Chemother. 1985 Jul;16 (Suppl A):63–90. doi: 10.1093/jac/16.suppl_a.63. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- Zalacain M., Cundliffe E. Methylation of 23S rRNA caused by tlrA (ermSF), a tylosin resistance determinant from Streptomyces fradiae. J Bacteriol. 1989 Aug;171(8):4254–4260. doi: 10.1128/jb.171.8.4254-4260.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]






