Abstract
Traditional and molecular typing schemes for the characterization of pathogenic microorganisms are poorly portable because they index variation that is difficult to compare among laboratories. To overcome these problems, we propose multilocus sequence typing (MLST), which exploits the unambiguous nature and electronic portability of nucleotide sequence data for the characterization of microorganisms. To evaluate MLST, we determined the sequences of ≈470-bp fragments from 11 housekeeping genes in a reference set of 107 isolates of Neisseria meningitidis from invasive disease and healthy carriers. For each locus, alleles were assigned arbitrary numbers and dendrograms were constructed from the pairwise differences in multilocus allelic profiles by cluster analysis. The strain associations obtained were consistent with clonal groupings previously determined by multilocus enzyme electrophoresis. A subset of six gene fragments was chosen that retained the resolution and congruence achieved by using all 11 loci. Most isolates from hyper-virulent lineages of serogroups A, B, and C meningococci were identical for all loci or differed from the majority type at only a single locus. MLST using six loci therefore reliably identified the major meningococcal lineages associated with invasive disease. MLST can be applied to almost all bacterial species and other haploid organisms, including those that are difficult to cultivate. The overwhelming advantage of MLST over other molecular typing methods is that sequence data are truly portable between laboratories, permitting one expanding global database per species to be placed on a World-Wide Web site, thus enabling exchange of molecular typing data for global epidemiology via the Internet.
Keywords: molecular typing, Neisseria meningitidis, housekeeping genes, World-Wide Web, hyper-virulent clones
The ability to identify accurately the strains of infectious agents that cause disease is central to epidemiological surveillance and public health decisions, but there are no wholly satisfactory methods of achieving this goal (1). All of the numerous methods that are currently used suffer from significant drawbacks, including various combinations of inadequate discrimination, limited availability of reagents, poor reproducibility within and between laboratories, and an inability to quantitate the genetic relationships between isolates. However, perhaps the most important limitation of current typing methods is the difficulty of comparing the results achieved by different laboratories.
Molecular typing methods are used to address two very different kinds of problem. First, are the isolates recovered from a localized outbreak of disease the same or different strains (short term or local epidemiology)? Second, how are strains causing disease in one geographic area related to those isolated world-wide (long term or global epidemiology)? Different methods may be appropriate for investigating local and global epidemiology, but in both cases they should be highly discriminatory such that isolates assigned to the same molecular type are likely to be descended from a recent common ancestor, and isolates that share a more distant common ancestor are not assigned to the same type.
High levels of discrimination can be achieved in two quite different ways. In one approach, individual loci, or uncharacterized regions of the genome, that are highly variable within the bacterial population are identified. For bacterial pathogens, several methods based on this approach are currently popular, e.g., ribotyping, pulsed-field gel electrophoresis (PFGE), and PCR with repetitive element primers, or arbitrary primers (1). In these methods, restriction enzymes (or PCR primers) are chosen that give maximal variation within the population; consequently, the variation that is indexed is evolving very rapidly, usually for unknown reasons. The second approach, typified by multilocus enzyme electrophoresis (MLEE), is to use variation that is accumulating very slowly in the population and that is likely to be selectively neutral. Although only a small number of alleles can be identified within the population by using this type of variation, high levels of discrimination are achieved by analyzing many loci.
Methods that index rapidly evolving variation are useful for short term epidemiology but may be misleading for global epidemiology. Several studies have shown that techniques such as PFGE resolve isolates that are indistinguishable by MLEE. For example, MLEE studies of populations of Salmonella enterica have shown that isolates of serovar Typhi from typhoid fever belong to one of two closely related electrophoretic types (ETs) (2). In contrast, isolates of serovar Typhi are relatively diverse according to PFGE (3). PFGE is therefore useful for studying individual outbreaks of typhoid fever because, unlike MLEE, it identifies the microvariation that is needed to distinguish between strains circulating within a geographic area. However, this technique is too discriminatory for long term epidemiology because it does not indicate that isolates that cause typhoid fever are members of a single globally distributed clonal lineage of S. enterica. To use a common metaphor, PFGE and other similar methods fail to see the forest for the trees.
The most appropriate of the current techniques for long term epidemiology, and for the identification of lineages that have an increased propensity to cause disease, is undoubtedly MLEE. This approach also has contributed most to our understanding of the global epidemiology and population structure of infectious agents. For many pathogens, MLEE successfully has identified clusters of closely related strains (clones or clonal complexes) that are particularly liable to cause disease (1, 4). A major problem with MLEE, and all other current typing methods, is that the results obtained in different laboratories are difficult to compare. We have therefore chosen to adapt the proven concepts and methods of MLEE by identifying alleles directly from the nucleotide sequences of internal fragments of housekeeping genes rather than by comparing the electrophoretic mobilities of the enzymes they encode. This modification has overwhelming advantages. First, far more variation can be detected, resulting in many more alleles per locus than are obtained with MLEE. Second, sequence data can be compared readily between laboratories, such that a typing method based on the sequences of gene fragments from a number of different housekeeping loci [multilocus sequence typing (MLST)] is fully portable and data stored in a single expanding central multilocus sequence database can be interrogated electronically via the Internet to produce a powerful resource for global epidemiology. In this paper, we report the development and validation of MLST for the identification of the virulent lineages of the bacterial pathogen Neisseria meningitidis. The MLST approach is, however, applicable to almost all pathogenic, or nonpathogenic, bacterial species and to many other haploid organisms.
MATERIALS AND METHODS
Bacterial Strains.
A total of 107 strains of N. meningitidis were chosen for analysis from globally representative strain collections (5–8). The strains included ≈10 isolates of each of the 7 recognized hyper-virulent lineages (subgroups I, III, and IV-1, ET-5 complex, ET-37 complex, A4 cluster, and lineage 3), chosen to represent the diversity of MLEE profiles, dates, and countries of origin found within each lineage. One strain was chosen from each of the other serogroup A subgroups, and 30 strains were included to represent the diversity of the other ETs resolved by MLEE, most of which had been isolated in the Netherlands (9, 10) and Norway (11). Two strains (NG 3/88, NGH 41) that had been assigned to the A4 cluster on the basis of a dendrogram of serogroup B bacteria (8) had not clustered with the A4 cluster with data from a larger strain collection (5). They did not cluster with the A4 strains in this analysis and have been reassigned here as “other.”
Nucleotide Sequencing of Gene Fragments.
The nucleotide sequences of internal fragments of the following genes (protein products are shown in parentheses) were obtained: abcZ (putative ABC transporter), adk (adenylate kinase), aroE (shikimate dehydrogenase), gdh (glucose-6-phosphate dehydrogenase), mtg (monofunctional peptidoglycan transglycosylase), pdhC (pyruvate dehydrogenase subunit), pgm (phosphoglucomutase), pilA (regulator of pilin synthesis), pip (proline imino-peptidase), ppk (polyphosphate kinase), and serC (3-phosphoserine aminotransferase). The gene fragments were amplified from chromosomal DNA of the 107 N. meningitidis strains by using PCR with the following primers: abcZ-P1, 5′-AATCGTTTATGTACCGCAGG-3′ and abcZ-P2, 5′-GTTGATTTCTGCCTGTTCGG-3′; adk-P1, 5′-ATGGCAGTTTGTGCAGTTGG-3′ and adk-P2, 5′-GATTTAAACAGCGATTGCCC-3′; aroE-P1, 5′-ACGCATTTGCGCCGACA- TC-3′ and aroE-P2, 5′-ATCAGGGCTTTTTTCAGGTT-3′; gdh-P1, 5′-ATCAATACCGATGTGGCGCGT-3′ and gdh-P2, 5′-GGTTTTCATCTGCGTATAGAG-3′; mtg-P1, 5′-CGGCATCTTTATCTTTTTCAA-3′ and mtg-P2, 5′-TCAGTCCGTA/GTCNCTT/CTCNGG-3′; pdhC-P1, 5′-GGTTTCCAACGTATCGGCGAC-3′ and pdhC-P2, 5′-ATCGGCTTTGATGCCGTATTT-3′; pgm-P1, 5′-CTTCAAAGCCTACGACATCCG-3′ and pgm-P2, 5′-CGGATTGCTTTCGATGACGGC-3′; pilA-P1, 5′-AAGGGCTGAAAGACGGCAA-3′ and pilA-P2, 5′-CAATCCAGCAGTCGGTCCACA-3′; pip-P1, 5′-CGGATACTTGCAGGTGTCTG-3′ and pip-P2, 5′-CTCAACCGCCTGAACCAACG-3′; ppk-P1 5′-GAACAAAACCGCATCCTCTGC-3′ and ppk-P2, 5′-ATCGTTTTGCAGGTCGGCTTC-3′; and serC-P1, 5′ CTGCCAGCCTAAAATCGGGCGGGTTATTG-3′ and serC-P2, 5′-CAACATCGGGACATCAACCG-3′. Sequencing of both strands of the amplified fragments was achieved by using an Applied Biosystems Prism 377 automated sequencer with dRhodamine-labeled terminators (PE Applied Biosystems). The following primers were used for sequencing: abcZ-P1 and abcZ-S2, 5′-GAGAACGAGCCGGGATAGGA-3′; adk-S1, 5′-AGGCTGGCACGCCCTTGG-3′ and adk-S2, 5′-CAATACTTCGGCTTTCACGG-3′; aroE-S1, 5′-GCGGTCAAC/TACGCTGATT-3′ and aroE-S2, 5′-ATGATGTTGCCGTACACATA-3′; gdh-S1, 5′-GTGGCGCGTTATTTCAAAGA-3′ and gdh-S2, 5′-CTGCCTTCAAAAATATGGCT-3′; mtg-S1, 5′-CTATGTGTACGGCAACATCAT-3′ and mtg-P2; pdhC-S1, 5′- TCTACTACATCACCCTGATG-3′ and pdhC-P2; pgm-S1, 5′-CGGCGATGCCGACCGCTTGG-3′ and pgm-S2, 5′-GGTGATGATTTCGGTTGCGCC-3′; pilA-P1 and pilA-S2, 5′-GGCTTTGACTTGGTTGACGG-3′; pip-P1 and pip-S2, 5′-GATTTTCAGCAATCGGCGCG-3′; ppk-P1 and ppk-S2, 5′-GGCAGCCTTTGACGTTCATGC-3′; and serC-S1, 5′-CAACGGGCTGCAATACCGTG-3′ and serC-P2.
Chromosomal Mapping.
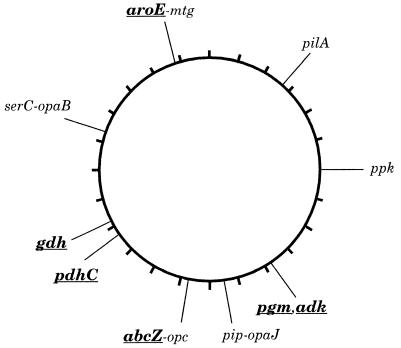
Gene fragments were amplified as above by using the PCR digoxygenin labeling mix (Boehringer Mannheim) and hybridized to chromosomal DNA from strain Z2491 (subgroup IV-1), which had been separated by PFGE after digestion with the rare cutting enzymes SgfI, NheI, SpeI, BglII, PmeI, or PacI. The bands that hybridized were identified on the physical map of strain Z2491 (12). The data confirmed the published map locations of pgm, ppk, and pdhC (12). serC maps near opaB (13), and abcZ maps near opc (data not shown), whose map locations also were confirmed. The map locations of these and the newly mapped gene fragments gdh, aroE-mtg, pilA, adk, and pip are shown in Fig. 1.
Figure 1.
Chromosomal locations of gene fragments. The locations are drawn on the physical map of strain Z2491 (12), a subgroup IV-1 strain. The six loci chosen for MLST are shown in boldfaced, underlined text. aroE and mtg are located next to each other (14) on BglII fragment B14 (41 kb). pip and opaJ are also next to each other (13) (data not shown) and are located on BglII fragment B16 (32 kb). serC and opaB are located within a few kilobases of each other (13) as are abcZ and opc (unpublished data). pgm and adk hybridized to the same set of fragments, including B7 and P3, which overlap by ≈50 kb. gdh mapped on SpeI fragment S17 (35 kb).
Estimating Relatedness Between Strains.
For each gene fragment, the sequences from the 107 strains were compared, and isolates with identical sequences were assigned the same allele number. For each strain, the combination of alleles at each locus defined its multilocus sequence type (ST). The relatedness between each ST was shown as a dendrogram, constructed by the unweighted pair group cluster method with arithmetic averages [unweighted pair group method with arithmetic mean (UPGMA)] from the matrix of allelic mismatches between the STs.
RESULTS
The Population Structure of N. meningitidis.
N. meningitidis, the meningococcus, is a major bacterial pathogen that causes epidemics, outbreaks, and isolated cases of meningitis and septicemia globally. We chose this species to validate MLST because a set of reference strains was available whose relationships have been inferred by using MLEE. In addition, meningococci present a particular challenge to molecular typing because the extent of recombination in meningococci is higher than that in most bacterial populations (15).
Populations of bacterial pathogens typically consist of a large and heterogeneous collection of isolates that rarely cause disease and a small number of groups of closely related strains (clones or lineages) that are particularly associated with outbreaks of disease. We will use the term “hyper-virulent lineage” to describe strains with an increased capacity to cause disease. Most invasive meningococcal disease in the developed world has been associated with a small number of hyper-virulent lineages of serogroup B or C isolates, referred to by MLEE designations: ET-5 complex; ET-37 complex; A4 cluster; and lineage 3 (16). In parts of the developing world, and particularly in sub-Saharan Africa, epidemics or pandemics of meningococcal disease occur and usually are caused by isolates of serogroup A. Over the last 30 years, epidemics and pandemics of serogroup A meningococcal disease have been caused by a small number of related hyper-virulent lineages, termed “subgroups” (16), the most important of which are subgroups I, III, and IV-1.
Recombination in meningococci is believed to be frequent compared with mutation (17). Accordingly, hyper-virulent lineages will emerge at intervals within the population and slowly diversify as their initially uniform genomes become increasingly pocked by highly localized recombinational replacements. Ultimately, these lineages may diversify to such an extent that they can no longer be distinguished from the background meningococcal population. MLEE studies, using 12–19 loci, successfully have identified hyper-virulent lineages among meningococci as they form clusters (clone complexes) of closely related ETs on dendrograms constructed from the electrophoretic data (5–8).
Nucleotide sequencing of multiple housekeeping genes (possessing the appropriate levels of sequence diversity) should also assign strains to each of the known hyper-virulent lineages and distinguish these lineages from each other and from the large background of other isolates. Accordingly, all members of each of the currently circulating hyper-virulent lineages should have identical alleles at all housekeeping genes. Exceptions will occur where a recombinational replacement (or mutation) has occurred within one of the genes being sequenced. In contrast, most isolates from the general meningococcal population, e.g., those from the nasopharynges of healthy carriers, are known to be more diverse than disease isolates and will often have unique combinations of alleles at the housekeeping loci. The repeated isolation of meningococci that have the same alleles at each of the housekeeping loci identifies a hyper-virulent lineage or clone. The method therefore has the potential to identify existing and newly emerging hyper-virulent lineages and to monitor their global spread.
Sequences of Gene Fragments from 107 Reference Strains of N. meningitidis.
We chose internal regions of 11 housekeeping genes that were sufficiently small to be sequenced accurately using a single primer for each direction (417–579 bp). The sequences of these 11 gene fragments were determined for all 107 strains. The number of alleles ranged from 10 to 36, with 26–166 variable bases per gene fragment (Table 1). The genes were mapped on a physical map (12) to ensure that they were unlinked (Fig. 1). Of the 11 loci, only mtg and aroE were linked sufficiently closely that they might be frequently coinherited in single transformation events.
Table 1.
Gene fragments used in MLST analysis
| Gene | Fragment size, bp |
n
|
Anomalies*
|
||
|---|---|---|---|---|---|
| Alleles | Variable sites | MLEE | MLST | ||
| abcZ† | 433 | 15 | 75 | 3 (2) | 4 (3) |
| adk | 465 | 10 | 38 | 0 | 0 |
| aroE | 490 | 18 | 166 | 2 | 3 |
| gdh | 501 | 16 | 28 | 2 | 2 |
| mtg | 497 | 16 | 61 | 1 | 2 |
| pdhC | 480 | 24 | 80 | 2 | 2 |
| pgm | 450 | 21 | 77 | 3 | 3 |
| pilA | 432 | 36 | 50 | 12 (11) | 15 (14) |
| pip | 417 | 19 | 26 | 7 | 7 |
| ppk | 579 | 23 | 77 | 7 | 9 |
| serC | 451 | 29 | 67 | 13 (7) | 21 (15) |
The numbers of alleles in groups of at least four strains in the reference set of 107 meningococcal isolates that are inconsistent with strain relationships previously determined by MLEE or within the clustering obtained by MLST at a genetic distance of ≤0.35 in Fig. 1. The numbers in parentheses are the numbers of anomalous alleles when replicate anomalies within a given grouping are counted only once.
The six genes used to generate Fig. 2 are shown in boldface. These genes provided data that were consistent both between MLEE and MLST as well as within MLST groups. The mtg gene fragment was excluded from the set of six because of its close physical linkage to aroE.
Congruence Between MLST and MLEE.
We refer to a unique combination of alleles as a sequence type (ST), which is analogous to the MLEE electrophoretic type (ET). A dendrogram based on a matrix of pairwise differences between the allelic profiles for the 11 loci resolved 74 STs among the 107 strains and yielded results corresponding to those from MLEE (data not shown), with a few exceptions described below.
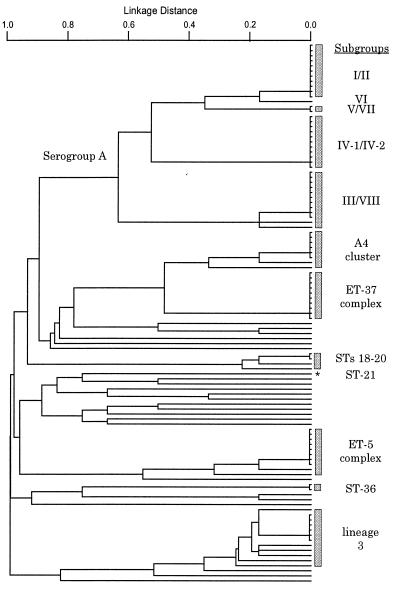
The congruence between sequence data and MLEE was much better for some gene fragments than others, for reasons that will be discussed elsewhere. We therefore chose a subset of six gene fragments (abcZ, adk, aroE, gdh, pdhC, and pgm) (Table 1) for which the allele assignments correlated almost perfectly with that expected from the MLEE data. Because this approach assumes the validity of the clustering produced by MLEE, the data also were analyzed for internal consistency that confirmed that these six loci were the most congruent (Table 1). The dendrogram constructed by using this subset of six loci (Fig. 2) was extremely similar to that obtained by using all 11 loci (data not shown) because the added resolution achieved by using more loci was counterbalanced by the decreased congruence obtained by using the extra loci. Isolates assigned by MLEE to the seven hyper-virulent lineages were distinguished clearly from each other and from other isolates (Fig. 2). For each of the seven hyper-virulent lineages, either all isolates tested were identical at all six loci (subgroups I and IV-1, ET-37 complex) or, with two exceptions, they differed from the most common ST at a single locus (subgroup III, A4 cluster, ET-5 complex, and lineage 3; Table 2). The two exceptions, one ET-5 complex isolate and one A4 cluster isolate, differed from the most common ST at two of the six loci.
Figure 2.
Dendrogram of genetic relationships among 107 strains based on 6 gene fragments. Linkage distance is indicated by a scale at the top, and the MLEE or ST assignments of lineages are indicated by shaded rectangles. The asterisk indicates ST-21 (serogroup A strain B534).
Table 2.
Properties of strains within 49 STs defined by alleles of six gene fragments
| ST | Strains, n | Reference strain | MLEE assignment | Continents | Years of isolation | Serogroup | Allele numbers
|
|||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| abcZ | pdhC | gdh | aroE | pgm | adk | |||||||
| 1 | 11 | B40 | Subgroups I, II | AF, AS, AU, EU, NA | 63-77 | A, C | 1 | 1 | 1 | 1 | 3 | 3 |
| 2 | 1 | Z4024 | Subgroup VI | EU | 85 | A | 1 | 1 | 1 | 4 | 3 | 3 |
| 3 | 2 | Z4081 | Subgroups V, VII | AS | 79 | A | 1 | 23 | 1 | 1 | 13 | 3 |
| 4 | 11 | Z2491 | Subgroups IV-1, IV-2 | AF, AS, NA | 37-90 | A | 1 | 2 | 4 | 3 | 3 | 3 |
| 5 | 10 | Z3524 | Subgroups III, VIII | AF, AS, EU, SA | 63-88 | A | 1 | 2 | 3 | 2 | 3 | 1 |
| 6 | 1 | Z3906 | Subgroup III | AS | 62 | A | 1 | 2 | 3 | 2 | 11 | 1 |
| 7 | 1 | Z5826 | Subgroup III | AS | 92 | A | 1 | 2 | 3 | 2 | 19 | 1 |
| 8 | 6 | BZ 10 | A4 cluster | AF, AS, EU | 67-92 | B, C | 2 | 5 | 8 | 7 | 2 | 3 |
| 9 | 1 | BZ 163 | A4 cluster | EU | 79 | B | 2 | 5 | 8 | 8 | 2 | 3 |
| 10 | 1 | B6116/77 | A4 cluster | EU | 77 | B | 2 | 15 | 8 | 4 | 2 | 3 |
| 11 | 10 | L93/4286 | ET-37 complex | AF, EU, NA, SA | 64-93 | B, C | 2 | 4 | 8 | 4 | 6 | 3 |
| 12 | 1 | NG 3/88 | Other | EU | 88 | B | 4 | 11 | 8 | 2 | 20 | 3 |
| 13 | 1 | NG 6/88 | Other | EU | 88 | B | 4 | 11 | 8 | 15 | 1 | 10 |
| 14 | 1 | NG F26 | Other | EU | 88 | B | 4 | 11 | 8 | 15 | 1 | 1 |
| 15 | 1 | NG E31 | Other | EU | 88 | B | 13 | 11 | 3 | 16 | 9 | 3 |
| 16 | 1 | DK 24 | Other | EU | 40 | B | 15 | 19 | 8 | 9 | 15 | 9 |
| 17 | 1 | 3906 | Other | AS | 77 | B | 8 | 12 | 11 | 13 | 4 | 3 |
| 18 | 2 | EG 328 | Other | EU | 85-89 | B | 7 | 1 | 10 | 10 | 2 | 8 |
| 19 | 1 | EG 327 | Other | EU | 85 | B | 7 | 1 | 8 | 10 | 2 | 8 |
| 20 | 1 | 1000 | Other | EU | 88 | B | 6 | 1 | 10 | 10 | 2 | 8 |
| 21 | 1 | B534 | Other | EU | 41 | A | 1 | 16 | 2 | 1 | 17 | 5 |
| 22 | 1 | A22 | Other | EU | 86 | W-135 | 11 | 24 | 11 | 18 | 21 | 5 |
| 23 | 1 | 71/94 | Other | EU | 94 | Y | 10 | 9 | 11 | 18 | 17 | 5 |
| 24 | 1 | 860060 | Other | EU | 86 | X | 2 | 20 | 15 | 2 | 5 | 5 |
| 25 | 1 | NG G40 | Other | EU | 88 | B | 6 | 13 | 6 | 2 | 14 | 5 |
| 26 | 1 | NG E28 | Other | EU | 88 | B | 6 | 10 | 12 | 2 | 14 | 5 |
| 27 | 1 | NG H41 | Other | EU | 88 | B | 3 | 18 | 7 | 6 | 2 | 5 |
| 28 | 1 | 890326 | Other | EU | 89 | Z | 13 | 18 | 5 | 6 | 2 | 4 |
| 29 | 1 | 860800 | Other | EU | 86 | Y | 2 | 18 | 16 | 6 | 8 | 7 |
| 30 | 1 | NG 4/88 | Other | EU | 88 | B | 6 | 21 | 1 | 6 | 8 | 5 |
| 31 | 1 | E32 | Other | EU | 88 | Z | 14 | 8 | 3 | 6 | 18 | 5 |
| 32 | 8 | 44/76 | ET-5 complex | EU, SA | 76-87 | B, C | 4 | 3 | 6 | 5 | 8 | 10 |
| 33 | 1 | 204/92 | ET-5 complex | NA | 92 | B | 8 | 3 | 6 | 5 | 8 | 10 |
| 34 | 1 | BZ 83 | ET-5 complex | EU | 84 | B | 8 | 3 | 5 | 5 | 8 | 10 |
| 35 | 1 | SWZ107 | Other | EU | 86 | B | 4 | 10 | 6 | 11 | 12 | 10 |
| 36 | 2 | NG H38 | Other | EU | 86-88 | B | 12 | 21 | 5 | 4 | 16 | 7 |
| 37 | 1 | DK 353 | Other | EU | 62 | B | 12 | 21 | 13 | 15 | 10 | 2 |
| 38 | 1 | BZ 232 | Other | EU | 64 | B | 12 | 17 | 13 | 15 | 10 | 2 |
| 39 | 1 | E26 | Other | EU | 88 | X | 5 | 7 | 14 | 17 | 16 | 4 |
| 40 | 1 | 400 | Lineage 3 | EU | 91 | B | 3 | 22 | 9 | 9 | 9 | 6 |
| 41 | 6 | BZ 198 | Lineage 3 | EU, SA | 86-96 | B | 3 | 6 | 9 | 9 | 9 | 6 |
| 42 | 1 | 91/40 | Lineage 3 | AS | 91 | B | 10 | 6 | 9 | 9 | 9 | 6 |
| 43 | 1 | NG H15 | Other | EU | 88 | B | 12 | 6 | 9 | 9 | 9 | 6 |
| 44 | 1 | NG E30 | Other | EU | 88 | B | 9 | 6 | 9 | 9 | 9 | 6 |
| 45 | 1 | 50/94 | Lineage 3 | EU | 94 | B | 3 | 6 | 9 | 9 | 15 | 6 |
| 46 | 1 | 88/03415 | Lineage 3 | EU | 88 | B | 3 | 6 | 3 | 9 | 9 | 6 |
| 47 | 1 | NG H36 | Other | EU | 88 | B | 9 | 6 | 9 | 9 | 2 | 6 |
| 48 | 1 | BZ 147 | Other | EU | 63 | B | 9 | 5 | 9 | 14 | 9 | 6 |
| 49 | 1 | 297-0 | Other | SA | 87 | B | 2 | 14 | 3 | 12 | 7 | 6 |
AF, Africa; AS, Asia including India; AU, Australia and New Zealand; EU, Europe including Iceland and Russia; NA, North America; SA, South America.
The serogroup A strains formed a cluster of lineages that were distinct from strains of other serogroups, with the exception of strain B534 (ST-21), and the major subgroups associated with epidemic meningitis (I, III, and IV-1) were distinguished easily (Fig. 2). The serogroup A strain B534 had been assigned to subgroup I by Wang et al. (7) but was not closely related to the other serogroup A strains by MLST. Recent MLEE data (D.A.C., unpublished data) support the MLST data and show that assignment of this strain to subgroup I was incorrect. MLST did not distinguish between isolates of serogroup A subgroups I and II, V and VII, IV-1 and IV-2, or III and VIII, but these four pairs of subgroups are known to be very closely related.
The A4 cluster and the ET-37 complex formed a cluster of lineages that were distinct from other STs. The ET-5 and lineage 3 strains each formed distinct clusters, although the lineage 3 strains were not well resolved from some unrelated STs.
Almost all of the isolates that had not been assigned to known hyper-virulent lineages by MLEE had unique unrelated STs. However, serogroup C strain BZ133 was identical to serogroup A subgroup I/II bacteria (ST-1). The MLEE profile of this strain was also indistinguishable from subgroup I strains, and it probably represents a subgroup I organism that has acquired a serogroup C capsule by transformation (18). Two strains (NG H15, ST-43; NGE 30, ST-44) clustered within lineage 3 according to the six gene fragments (Table 2). They were related to, but distinct from, lineage 3 when all 11 genes were compared and also differed from lineage 3 by MLEE. Additional sequence data from other conserved genes would be required to decide whether these two strains represent diverse variants of lineage 3 or not. ST-36 contained two isolates and STs 18–20 included four strains that clustered as closely together as did isolates belonging to the hyper-virulent lineages. These results suggest that additional lineages may exist that have not been documented extensively until now.
DISCUSSION
MLEE has provided an invaluable population genetic framework for bacterial and nonbacterial species and for the identification of clones that are particularly associated with disease (1, 4, 19, 20). However, MLEE relies on the indirect assignment of alleles based on the electrophoretic mobility of enzymes, and indistinguishable mobility variants may be encoded by very different nucleotide sequences. In MLST, the direct assignment of alleles based on nucleotide sequence determination of internal fragments from multiple housekeeping genes is unambiguous and distinguishes more alleles per locus, thus allowing high levels of discrimination between isolates by using half of the loci that are typically required for MLEE. For the six gene fragments chosen for typing meningococci, there was an average of 17 alleles per locus and the potential to resolve over 24 million STs. The use of multiple loci is essential to achieve the resolution required to provide meaningful relationships among strains and is particularly important because clones diversify with age, as a consequence of mutational or recombinational events, and might be typed incorrectly if only single loci were examined.
The relatively rapid diversification of clones by recombination was expected to be a significant problem with meningococci. However, the six loci chosen allow the reliable recognition of the isolates of the known hyper-virulent lineages. The members of each hyper-virulent lineage were identical at all six loci or differed from the consensus ST for that clone at only a single locus (with two exceptions) and were resolved on the dendrogram from the other major lineages. Furthermore, most of the other isolates were distinct from the hyper-virulent lineages, with the exception of some of the minor subgroups of serogroup A, and the strains that clustered among the lineage 3 strains. The inclusion of an additional highly congruent housekeeping gene may be required to improve the resolution of these strains.
MLEE is the currently accepted method for assigning meningococci to the known hyper-virulent lineages. We believe the advantages of MLST over MLEE for the characterization of meningococci are so considerable that we have set up a World-Wide Web site for MLST of meningococci (http://mlst.zoo.ox.ac.uk). Besides its portability, MLST has the advantage that it can be used after PCR amplification from clinical material (e.g., blood or cerebrospinal fluid), which is increasingly important because early provision of antibiotic treatment for meningitis results in bacteria being cultured less frequently. Although we stress that all six loci should be used to characterize meningococcal strains, it should be possible during investigations of outbreaks for public health purposes to determine rapidly whether the outbreak is caused by a single strain by using only two or three loci, and this data may provide a putative assignment to a known clonal lineage. Even with all six loci, assignment of a meningococcus to a known hyper-virulent lineage probably can be achieved at least as quickly and economically as by any currently available method.
MLST is a simple technique, requiring only the ability to amplify DNA fragments by PCR and to sequence the fragments, using an automated sequencer or manually. These techniques are, or will soon be, available to public health laboratories in the developed world and to an increasing number of laboratories in the developing world. Direct sequencing of ≈470-bp PCR products from hundreds of isolates currently can be carried out rapidly and accurately by using an automated DNA sequencer, and the complete assignment of alleles at six loci (sequencing on both DNA strands) can be accomplished by using only 12 lanes of a sequencing gel. Sequencing services also are being offered increasingly on a commercial basis, and the technology of automated sequencing is being improved rapidly.
The great advantage of MLST over MLEE and over molecular typing methods that rely on the comparisons of DNA fragment sizes is the unambiguity and portability of sequence data, which allow results from different laboratories to be compared without exchanging strains. This ability will allow laboratories in different countries and continents to relate their local isolates to those found globally by submitting the sequences from housekeeping gene fragments to a central World-Wide Web site containing the MLST database for that species. In addition, all of the components of an MLST analysis—genomic DNA, PCR products, and nucleotide sequencing reactions—are highly portable among laboratories by conventional mail, enabling typing to be carried out at remote sites and easy comparison of results among reference laboratories.
The sequence data obtained for MLST can be used to determine population structures by analyzing the extent of linkage disequilibrium between alleles and to look for recombination by the noncongruence of gene trees (21) and by the presence of significant mosaic structure (22). For highly clonal species, the phylogenetic relationships between isolates can be inferred from the dendrogram derived from the pairwise differences between STs and independently from a consensus tree constructed from the gene sequences. For weakly clonal species such as the meningococcus, MLST is very useful for the identification of the currently circulating hyper-virulent lineages because these are recognized as clusters of isolates with identical, or very similar, multilocus sequence types. Phylogenetic inferences from weakly clonal populations should be treated with caution, but the clustering of all serogroup A subgroups (Fig. 2) suggests that these may be descended from a common ancestor. Similarly, the close relationship between the A4 cluster and ET-37 complex suggests that they may be derived from a common ancestor. The population genetic inferences from the meningococcal data set will be discussed elsewhere.
We have chosen to develop and validate the utility of MLST by using N. meningitidis, a species that presents a particular challenge for typing methods, because of the rapid diversification of meningococcal clones by frequent localized recombinational exchanges among lineages. Because recombination did not prevent the identification of the hyper-virulent meningococcal clones, MLST should be suitable for almost any weakly clonal or clonal species with sufficient sequence diversity. MLST recently has been developed and validated for the identification of hyper-virulent clones of Streptococcus pneumoniae (M. C. Enright and B.G.S., unpublished work).
Currently, different typing methods often are used for the same pathogens in different laboratories and, even when a uniform method is used, the data are difficult to compare between laboratories and are often unsuitable for evolutionary, phylogenetic, or population genetic studies. Acceptance of MLST as the “gold standard” for typing bacterial pathogens would resolve this highly unsatisfactory situation. MLEE commonly is used for typing and population genetic analysis of pathogenic fungi and parasites, and MLST also should be useful for the determination of the population structures of nonbacterial haploid infectious agents and for portable molecular typing of those agents that are weakly or strongly clonal.
Acknowledgments
We thank Paul Wilkinson for his assistance. This work was supported from funds from the Wellcome Trust. M.C.J.M. is a Wellcome Trust Senior Research Fellow in Biodiversity. B.G.S. is a Wellcome Trust Principal Research Fellow. J.E.R. is supported by the Meningitis Research Foundation.
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviations: ET, electrophoretic type; MLST, multilocus sequence typing; MLEE, multilocus enzyme electrophoresis; PFGE, pulsed-field gel electrophoresis; ST, sequence type.
References
- 1.Achtman M. In: Molecular Medical Microbiology. Sussman M, editor. London: Academic; 1998. , in press. [Google Scholar]
- 2.Selander R K, Beltran P, Smith N H, Helmuth R, Rubin F A, Kopecko D J, Ferris K, Tall B T, Cravioto A, Musser J M. Infect Immun. 1990;58:2262–2275. doi: 10.1128/iai.58.7.2262-2275.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Navarro F, Llovet T, Echeita M A, Coll P, Aladueña A, Usera M A, Prats G. J Clin Microbiol. 1996;34:2831–2834. doi: 10.1128/jcm.34.11.2831-2834.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Selander R K, Musser J M, Caugant D A, Gilmour M N, Whittam T S. Microb Pathog. 1987;3:1–7. doi: 10.1016/0882-4010(87)90032-5. [DOI] [PubMed] [Google Scholar]
- 5.Caugant D A, Bøvre K, Gaustad P, Bryn K, Holten E, Høiby E A, Frøholm L O. J Gen Microbiol. 1986;132:641–652. doi: 10.1099/00221287-132-3-641. [DOI] [PubMed] [Google Scholar]
- 6.Wang J, Caugant D A, Morelli G, Koumaré B, Achtman M. J Infect Dis. 1993;167:1320–1329. doi: 10.1093/infdis/167.6.1320. [DOI] [PubMed] [Google Scholar]
- 7.Wang J, Caugant D A, Li X, Hu X, Poolman J T, Crowe B A, Achtman M. Infect Immun. 1992;60:5267–5282. doi: 10.1128/iai.60.12.5267-5282.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Seiler A, Reinhardt R, Sarkari J, Caugant D A, Achtman M. Mol Microbiol. 1996;19:841–856. doi: 10.1046/j.1365-2958.1996.437970.x. [DOI] [PubMed] [Google Scholar]
- 9.Scholten R J P M, Poolman J T, Valkenburg H A, Bijlmer H A, Dankert J, Caugant D A. J Infect Dis. 1994;169:673–676. doi: 10.1093/infdis/169.3.673. [DOI] [PubMed] [Google Scholar]
- 10.Scholten R J P M, Bijlmer H A, Poolman J T, Kuipers B, Caugant D A, van Alphen L, Dankert J, Valkenburg H A. J Infect Dis. 1993;16:237–246. doi: 10.1093/clind/16.2.237. [DOI] [PubMed] [Google Scholar]
- 11.Caugant D A, Høiby E A, Magnus P, Scheel O, Hoel T, Bjune G, Wedege E, Eng J, Frøholm L O. J Clin Microbiol. 1994;32:323–330. doi: 10.1128/jcm.32.2.323-330.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dempsey J A F, Wallace A B, Cannon J G. J Bacteriol. 1995;177:6390–6400. doi: 10.1128/jb.177.22.6390-6400.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morelli G, Malorny B, Müller K, Seiler A, Wang J, del Valle J, Achtman M. Mol Microbiol. 1997;25:1047–1064. doi: 10.1046/j.1365-2958.1997.5211882.x. [DOI] [PubMed] [Google Scholar]
- 14.Zhou J J, Bowler L D, Spratt B G. Mol Microbiol. 1997;23:799–812. doi: 10.1046/j.1365-2958.1997.2681633.x. [DOI] [PubMed] [Google Scholar]
- 15.Spratt B G, Smith N H, Zhou J, O’Rourke M, Feil E. In: The Population Genetics of the Pathogenic Neisseria. Baumberg S, Young J P W, Saunders J R, Wellington E M H, editors. Cambridge, U.K.: Cambridge Univ. Press; 1995. pp. 143–160. [Google Scholar]
- 16.Achtman M. In: Meningococcal Disease. Cartwright K, editor. New York: Wiley; 1995. pp. 159–175. [Google Scholar]
- 17.Maiden M C J, Malorny B, Achtman M. Mol Microbiol. 1996;21:1297–1298. doi: 10.1046/j.1365-2958.1996.981457.x. [DOI] [PubMed] [Google Scholar]
- 18.Swartley J S, Marfin A A, Edupuganti S, Liu L J, Cieslak P, Perkins B, Wenger J D, Stephens D S. Proc Natl Acad Sci USA. 1997;94:271–276. doi: 10.1073/pnas.94.1.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tibayrenc M. Annu Rev Microbiol. 1996;50:401–429. doi: 10.1146/annurev.micro.50.1.401. [DOI] [PubMed] [Google Scholar]
- 20.Spratt B G, Feil E, Smith N H. In: Molecular Medical Microbiology. Sussman M, editor. London: Academic; 1998. , in press. [Google Scholar]
- 21.Boyd E F, Wang F-S, Whittam T S, Selander R K. Appl Environ Microbiol. 1996;62:804–808. doi: 10.1128/aem.62.3.804-808.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maynard Smith J. J Mol Evol. 1992;34:126–129. doi: 10.1007/BF00182389. [DOI] [PubMed] [Google Scholar]