Abstract
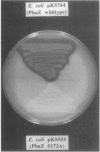
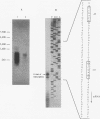
phaZPfi, the gene encoding the extracellular poly(3-hydroxyoctanoic acid) depolymerase of Pseudomonas fluorescens GK13, was cloned, sequenced, and characterized. It comprises 837 bp and is transcribed as a monocistronic message of about 950 bp from a putative sigma 70-like promoter 32 bp upstream of the ATG start codon. The deduced protein of 278 amino acids reveals a typical leader peptide at its N terminus. When expressed in Escherichia coli, the mature depolymerase started with Ala-23, whereas the mature enzyme purified from P. fluorescens GK13 started with both Leu-34 and Arg-35 determining proteins of 26,687 and 26,573 Da, respectively. The depolymerase is a strongly hydrophobic protein and includes the lipase consensus sequence Gly-X-Ser-X-Gly, which is known for serine hydrolases. Replacement of the central residue, Ser-172, in the corresponding sequence (Gly-Ile-Ser-Ser-Gly) of PhaZPfl with alanine resulted in complete loss of enzyme activity, indicating that the poly(3-hydroxyoctanoic acid) depolymerase belongs to the family of serine hydrolases.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson A. J., Dawes E. A. Occurrence, metabolism, metabolic role, and industrial uses of bacterial polyhydroxyalkanoates. Microbiol Rev. 1990 Dec;54(4):450–472. doi: 10.1128/mr.54.4.450-472.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Briese B. H., Jendrossek D., Schlegel H. G. Degradation of poly(3-hydroxybutyrate-co-3-hydroxyvalerate) by aerobic sewage sludge. FEMS Microbiol Lett. 1994 Mar 15;117(1):107–111. doi: 10.1111/j.1574-6968.1994.tb06750.x. [DOI] [PubMed] [Google Scholar]
- Brucato C. L., Wong S. S. Extracellular poly(3-hydroxybutyrate) depolymerase from Penicillium funiculosum: general characteristics and active site studies. Arch Biochem Biophys. 1991 Nov 1;290(2):497–502. doi: 10.1016/0003-9861(91)90572-z. [DOI] [PubMed] [Google Scholar]
- Budwill K., Fedorak P. M., Page W. J. Methanogenic degradation of poly(3-hydroxyalkanoates). Appl Environ Microbiol. 1992 Apr;58(4):1398–1401. doi: 10.1128/aem.58.4.1398-1401.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CHOWDHURY A. A. POLY-BETA-HYDROXYBUTTERSAEURE ABBAUENDE BAKTERIEN UND EXOENZYM. Arch Mikrobiol. 1963 Dec 10;47:167–200. [PubMed] [Google Scholar]
- Dalbey R. E. Leader peptidase. Mol Microbiol. 1991 Dec;5(12):2855–2860. doi: 10.1111/j.1365-2958.1991.tb01844.x. [DOI] [PubMed] [Google Scholar]
- Delafield F. P., Doudoroff M., Palleroni N. J., Lusty C. J., Contopoulos R. Decomposition of poly-beta-hydroxybutyrate by pseudomonads. J Bacteriol. 1965 Nov;90(5):1455–1466. doi: 10.1128/jb.90.5.1455-1466.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deretic V., Schurr M. J., Boucher J. C., Martin D. W. Conversion of Pseudomonas aeruginosa to mucoidy in cystic fibrosis: environmental stress and regulation of bacterial virulence by alternative sigma factors. J Bacteriol. 1994 May;176(10):2773–2780. doi: 10.1128/jb.176.10.2773-2780.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FORSYTH W. G., HAYWARD A. C., ROBERTS J. B. Occurrence of poly-beta-hydroxybutyric acid in aerobic gram-negative bacteria. Nature. 1958 Sep 20;182(4638):800–801. doi: 10.1038/182800a0. [DOI] [PubMed] [Google Scholar]
- Foster L. J., Lenz R. W., Fuller R. C. Quantitative determination of intracellular depolymerase activity in Pseudomonas oleovorans inclusions containing poly-3-hydroxyalkanoates with long alkyl substituents. FEMS Microbiol Lett. 1994 May 15;118(3):279–282. doi: 10.1111/j.1574-6968.1994.tb06841.x. [DOI] [PubMed] [Google Scholar]
- Frenken L. G., Egmond M. R., Batenburg A. M., Bos J. W., Visser C., Verrips C. T. Cloning of the Pseudomonas glumae lipase gene and determination of the active site residues. Appl Environ Microbiol. 1992 Dec;58(12):3787–3791. doi: 10.1128/aem.58.12.3787-3791.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gierasch L. M. Signal sequences. Biochemistry. 1989 Feb 7;28(3):923–930. doi: 10.1021/bi00429a001. [DOI] [PubMed] [Google Scholar]
- Grunstein M., Wallis J. Colony hybridization. Methods Enzymol. 1979;68:379–389. doi: 10.1016/0076-6879(79)68027-8. [DOI] [PubMed] [Google Scholar]
- Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983 Jun 5;166(4):557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- Haywood G. W., Anderson A. J., Ewing D. F., Dawes E. A. Accumulation of a Polyhydroxyalkanoate Containing Primarily 3-Hydroxydecanoate from Simple Carbohydrate Substrates by Pseudomonas sp. Strain NCIMB 40135. Appl Environ Microbiol. 1990 Nov;56(11):3354–3359. doi: 10.1128/aem.56.11.3354-3359.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huisman G. W., Wonink E., Meima R., Kazemier B., Terpstra P., Witholt B. Metabolism of poly(3-hydroxyalkanoates) (PHAs) by Pseudomonas oleovorans. Identification and sequences of genes and function of the encoded proteins in the synthesis and degradation of PHA. J Biol Chem. 1991 Feb 5;266(4):2191–2198. [PubMed] [Google Scholar]
- Huisman G. W., de Leeuw O., Eggink G., Witholt B. Synthesis of poly-3-hydroxyalkanoates is a common feature of fluorescent pseudomonads. Appl Environ Microbiol. 1989 Aug;55(8):1949–1954. doi: 10.1128/aem.55.8.1949-1954.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaeger K. E., Ransac S., Koch H. B., Ferrato F., Dijkstra B. W. Topological characterization and modeling of the 3D structure of lipase from Pseudomonas aeruginosa. FEBS Lett. 1993 Oct 11;332(1-2):143–149. doi: 10.1016/0014-5793(93)80501-k. [DOI] [PubMed] [Google Scholar]
- Jendrossek D., Müller B., Schlegel H. G. Cloning and characterization of the poly(hydroxyalkanoic acid)-depolymerase gene locus, phaZ1, of Pseudomonas lemoignei and its gene product. Eur J Biochem. 1993 Dec 1;218(2):701–710. doi: 10.1111/j.1432-1033.1993.tb18424.x. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A., Bebenek K., McClary J. Efficient site-directed mutagenesis using uracil-containing DNA. Methods Enzymol. 1991;204:125–139. doi: 10.1016/0076-6879(91)04008-c. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A., Roberts J. D., Zakour R. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lonetto M. A., Brown K. L., Rudd K. E., Buttner M. J. Analysis of the Streptomyces coelicolor sigE gene reveals the existence of a subfamily of eubacterial RNA polymerase sigma factors involved in the regulation of extracytoplasmic functions. Proc Natl Acad Sci U S A. 1994 Aug 2;91(16):7573–7577. doi: 10.1073/pnas.91.16.7573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mergaert J., Webb A., Anderson C., Wouters A., Swings J. Microbial degradation of poly(3-hydroxybutyrate) and poly(3-hydroxybutyrate-co-3-hydroxyvalerate) in soils. Appl Environ Microbiol. 1993 Oct;59(10):3233–3238. doi: 10.1128/aem.59.10.3233-3238.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama K., Saito T., Fukui T., Shirakura Y., Tomita K. Purification and properties of extracellular poly(3-hydroxybutyrate) depolymerases from Pseudomonas lemoignei. Biochim Biophys Acta. 1985 Jan 21;827(1):63–72. doi: 10.1016/0167-4838(85)90101-3. [DOI] [PubMed] [Google Scholar]
- Neu H. C., Heppel L. A. The release of enzymes from Escherichia coli by osmotic shock and during the formation of spheroplasts. J Biol Chem. 1965 Sep;240(9):3685–3692. [PubMed] [Google Scholar]
- Persson B., Bengtsson-Olivecrona G., Enerbäck S., Olivecrona T., Jörnvall H. Structural features of lipoprotein lipase. Lipase family relationships, binding interactions, non-equivalence of lipase cofactors, vitellogenin similarities and functional subdivision of lipoprotein lipase. Eur J Biochem. 1989 Jan 15;179(1):39–45. doi: 10.1111/j.1432-1033.1989.tb14518.x. [DOI] [PubMed] [Google Scholar]
- Pugsley A. P. The complete general secretory pathway in gram-negative bacteria. Microbiol Rev. 1993 Mar;57(1):50–108. doi: 10.1128/mr.57.1.50-108.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SCHLEGEL H. G., KALTWASSER H., GOTTSCHALK G. [A submersion method for culture of hydrogen-oxidizing bacteria: growth physiological studies]. Arch Mikrobiol. 1961;38:209–222. [PubMed] [Google Scholar]
- Saito T., Suzuki K., Yamamoto J., Fukui T., Miwa K., Tomita K., Nakanishi S., Odani S., Suzuki J., Ishikawa K. Cloning, nucleotide sequence, and expression in Escherichia coli of the gene for poly(3-hydroxybutyrate) depolymerase from Alcaligenes faecalis. J Bacteriol. 1989 Jan;171(1):184–189. doi: 10.1128/jb.171.1.184-189.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirmer A., Jendrossek D., Schlegel H. G. Degradation of poly(3-hydroxyoctanoic acid) [P(3HO)] by bacteria: purification and properties of a P(3HO) depolymerase from Pseudomonas fluorescens GK13. Appl Environ Microbiol. 1993 Apr;59(4):1220–1227. doi: 10.1128/aem.59.4.1220-1227.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinbüchel A., Hustede E., Liebergesell M., Pieper U., Timm A., Valentin H. Molecular basis for biosynthesis and accumulation of polyhydroxyalkanoic acids in bacteria. FEMS Microbiol Rev. 1992 Dec;9(2-4):217–230. doi: 10.1111/j.1574-6968.1992.tb05841.x. [DOI] [PubMed] [Google Scholar]
- Sutcliffe J. G. Nucleotide sequence of the ampicillin resistance gene of Escherichia coli plasmid pBR322. Proc Natl Acad Sci U S A. 1978 Aug;75(8):3737–3741. doi: 10.1073/pnas.75.8.3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanio T., Fukui T., Shirakura Y., Saito T., Tomita K., Kaiho T., Masamune S. An extracellular poly(3-hydroxybutyrate) depolymerase from Alcaligenes faecalis. Eur J Biochem. 1982 May;124(1):71–77. doi: 10.1111/j.1432-1033.1982.tb05907.x. [DOI] [PubMed] [Google Scholar]
- Timm A., Steinbüchel A. Cloning and molecular analysis of the poly(3-hydroxyalkanoic acid) gene locus of Pseudomonas aeruginosa PAO1. Eur J Biochem. 1992 Oct 1;209(1):15–30. doi: 10.1111/j.1432-1033.1992.tb17256.x. [DOI] [PubMed] [Google Scholar]
- Timm A., Steinbüchel A. Formation of polyesters consisting of medium-chain-length 3-hydroxyalkanoic acids from gluconate by Pseudomonas aeruginosa and other fluorescent pseudomonads. Appl Environ Microbiol. 1990 Nov;56(11):3360–3367. doi: 10.1128/aem.56.11.3360-3367.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tommassen J., Filloux A., Bally M., Murgier M., Lazdunski A. Protein secretion in Pseudomonas aeruginosa. FEMS Microbiol Rev. 1992 Sep;9(1):73–90. doi: 10.1016/0378-1097(92)90336-m. [DOI] [PubMed] [Google Scholar]
- Vieira J., Messing J. Production of single-stranded plasmid DNA. Methods Enzymol. 1987;153:3–11. doi: 10.1016/0076-6879(87)53044-0. [DOI] [PubMed] [Google Scholar]
- West S. E., Iglewski B. H. Codon usage in Pseudomonas aeruginosa. Nucleic Acids Res. 1988 Oct 11;16(19):9323–9335. doi: 10.1093/nar/16.19.9323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worsey M. J., Williams P. A. Metabolism of toluene and xylenes by Pseudomonas (putida (arvilla) mt-2: evidence for a new function of the TOL plasmid. J Bacteriol. 1975 Oct;124(1):7–13. doi: 10.1128/jb.124.1.7-13.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada K., Mukai K., Doi Y. Enzymatic degradation of poly(hydroxyalkanoates) by Pseudomonas pickettii. Int J Biol Macromol. 1993 Aug;15(4):215–220. doi: 10.1016/0141-8130(93)90040-s. [DOI] [PubMed] [Google Scholar]
- de Smet M. J., Eggink G., Witholt B., Kingma J., Wynberg H. Characterization of intracellular inclusions formed by Pseudomonas oleovorans during growth on octane. J Bacteriol. 1983 May;154(2):870–878. doi: 10.1128/jb.154.2.870-878.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Heijne G. Patterns of amino acids near signal-sequence cleavage sites. Eur J Biochem. 1983 Jun 1;133(1):17–21. doi: 10.1111/j.1432-1033.1983.tb07424.x. [DOI] [PubMed] [Google Scholar]